5627
3D isotropic multi-parameter mapping and synthetic imaging of the brain with 3D-QALAS: comparison with 2D MAGIC1Department of Imaging Physics, The University of Texas M.D. Anderson Cancer Center, Houston, TX, United States, 2MR Applications and Workflow, GE Healthcare, Menlo Park, CA, United States, 3MR Applications and Workflow, GE Healthcare, Waukesha, WI, United States, 4SyntheticMR, Linkoping, Sweden
Synopsis
3D QALAS is a promising new technique that simultaneously maps T1, T2, and PD in a single 3D acquisition. Both 3D QALAS and 2D MAGIC were applied for synthetic imaging and quantification in human brains. While T1 and T2 values were comparable between the two techniques, 3D QALAS was simpler to process and achieved smaller voxel sizes over larger matrices with similar acquisition times, resulting in less partial volume effects. 2D MAGIC maintained the high in-plane resolution and efficiency of an interleaved multi-slice technique. 3D QALAS thus presents an attractive quantification method for therapy planning and tissue volume measurement applications.
Introduction
The introduction of SyMRI and MAGIC [1] have enabled rapid multi-parameter mapping and synthetic imaging for clinical use. This is a 2D multi-slice technique that compares well with conventional 2D imaging in terms of contrast, resolution, and slice geometry. 3D QALAS [2] (3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse) is a more recent development that potentially provides the same capabilities in a 3D format, enabling isotropic imaging with smaller voxel sizes. While 3D QALAS was originally developed for breathhold cardiac imaging, in this work we adapt it for brain imaging and evaluate it with regards to several key factors, including resolution, efficiency, parameter fitting and accuracy, and clinical utility. These data are compared with those acquired with the 2D MAGIC from the same subject.Methods
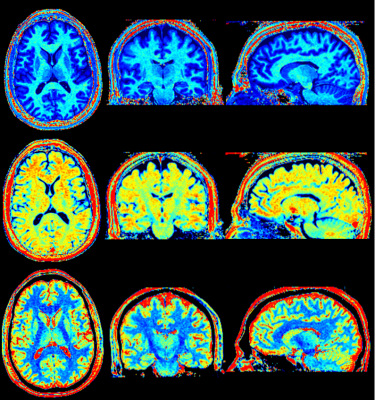
The 3D QALAS sequence was implemented for untriggered acquisitions in the head and body. This sequence consists of a 5 phases of varying contrasts acquired with 3D gradient echo readouts in a single cycled acquisition. A T2 prep sequence precedes the first phase, followed by an inversion prep pulse and 4 delayed phases (figure 1). One cycle would acquire one segment of Cartesian k-space for each of the 5 phases with spoiled gradient echo readouts, such that repeating the cycle would fill all segments of k-space for 5 image sets. The T2 prep time and the initial delay after the inversion prep pulse were both set at 100 msec. The sequence timing was arranged such that the 5 phases of the acquisition were equally spaced in time at 0.87-1.0 sec, mimicking a triggered cardiac acquisition. Parameter optimization was performed for isotropic voxel 3D axial slab brain imaging, while targeting a total scan time 6:00-6:30.
3D QALAS data was acquired of the brain in 5 healthy volunteers on a 3T scanner (MR750, GE Healthcare, Waukesha, WI). 2D MAGIC was also acquired with FOV=24.0, Matrix = 320x256, slice thickness/skip = 4/1 mm, TR = 5000, BW = +/- 20.83. Quantitative T1, T2, and PD maps were reconstructed from both acquisitions using a research version of SyMRI (SyntheticMR, Linkoping, Sweden) and an estimated B1 correction map was generated from the 2D data. Synthetic T1, T2, and FLAIR images were also reconstructed from the resulting maps. Volume segmentation of grey matter, white matter, and CSF was also performed based on grouped voxels of corresponding T1 and T2 values on the spin parameter maps.
For each subject, parameter maps and synthetic images were compared between the two acquisition types. 3D data was reformatted and viewed in different planes, and 3D and 2D data were compared for visual similarity and effects of partial volume at tissue interfaces.
Results
Sequence parameters for brain imaging were: FOV = 24.0-25.6 cm, matrix = 224x244, slice locations = 128, thickness = 1.2-1.3, flip = 4, views per segment=100, acceleration factor = 2. This resulted in isotropic resolution of 1.2-1.3 mm with whole brain coverage. Axial parameter maps were visually similar between the two techniques and provided measurements corresponding to reported values for brain tissues. Isotropic 3D maps and images could be viewed at any orthogonal plane without loss of resolution (figures 2 and 3).Discussion and Conclusion
From our experience with 3D multi-parameter mapping, we could identify several advantages of the technique. The smaller voxel sizes of the 3D acquisition reduce the effect of partial volume for parameter fitting, which can be a challenge at the interfaces of tissues with widely disparate T1 and T2 values. While a B1 measurement was not built into the 3D technique, the effects of B1 inhomogeneity were relatively small over the entire brain volume, as observed by other groups using modeled 3D techniques [3]. It also does not require modeling of the slice profile for much of the center portion of the slab. The isotropic acquisition provided less discretized data for volume measurements of segmented tissue. It could also be visualized at any angle, providing additional perspective or context for diagnostic evaluation. On the other hand, 2D MAGIC demonstrated better efficiency for axial in-plane viewing. It also had excellent SNR for the acquired resolution and scan time due to interleaved fast spin echo acquisition. While the 3D technique was acquired on a 3D Cartesian grid, it opens up the potential for use with compressed sensing or 3D radial acquisitions as well. Thus 3D QALAS is an excellent technique for multiplanar diagnosis, as well as for therapy planning and tissue volume segmentation applications.Acknowledgements
Research support was provided in part by GE Healthcare.References
1. Warntjes JB, Leinhard OD, West J, Lundberg P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn Reson Med. 2008 ; 60:320-9.
2. Kvernby S, Warntjes MJ, Haraldsson H, Carlhäll CJ, Engvall J, Ebbers T. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. J Cardiovasc Magn Reson. 2014; 16:102.
3. Tamir JI, Uecker M, Chen W, Lai P, Alley MT, Vasanawala SS, Lustig M. T2 shuffling: Sharp, multicontrast, volumetric fast spin-echo imaging. Magn Reson Med. 2017; 77:180-195.
Figures
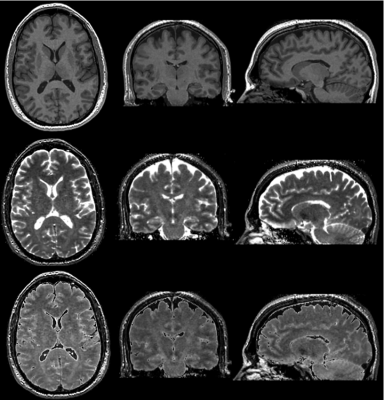
3D reformats of synthetic T1 weighted (top), T2 weighted (middle), and FLAIR (bottom) images.