5608
Magnitude and Complex Single- and Multi-echo Water Fat Separation via End-to-End Deep Learning1Research and Education, St Francis Hospital, Roslyn, NY, United States
Synopsis
The feasibility of water-fat separation using an end-to-end ConvNet approach was demonstrated for complex, magnitude and single echo acquisitions. The ConvNet approach showed images visually comparable to the GraphCut method with slightly higher signal to noise in typical cardiac image planes. Quantitative PDFF, R2* and off-resonance values had excellent correlation with a conventional analytical model based method. ConvNet based water-fat separation is a promising method capable of learning the water-fat separation problem with corrections for bipolar gradients, a multi-peak model, R2* and off-resonance.
Purpose
A new method for magnetic resonance (MR) imaging water-fat separation using a convolutional neural network (ConvNet) and deep learning (DL) is presented. Feasibility of the method with complex and magnitude images is demonstrated with a series of patient studies and accuracy of predicted quantitative values is analyzed.Methods
This retrospective study used complex raw data from a database of prospective research cardiovascular studies. The database contained relevant studies from 66 subjects: 17 subjects without a cardiovascular history or complaints (normal controls), 13 acute myocardial infarction (MI) subjects at 3 days after infarction and 34 chronic MI subjects (>2 years after infarction). Additionally, there were 26 repeat sessions with the acute MI patients in the subacute phase of infarction. In total, 90 imaging session were included. The database consisted of 1204 acquisitions.
All MR examinations were performed using one of two clinical 1.5 T imagers. The pulse sequence was a dark blood double inversion recovery multiple spoiled gradient-echo sequence (1 slice per breathhold, repetition time = 20 ms; 12 echo times, 2.4 - 15.5 ms (1.2 ms spacing), flip angle = 20 degrees, bandwidth = 1860 Hz/pixel, in-plane spatial resolution = 2.3 x 1.7 mm, slice thickness = 8 mm, flow compensation in read and slice). Every subject had acquisitions in 2-, 3- and 4- chamber long axis as well as short axis planes spanning the left ventricle. Multi-channel complex raw data was saved to the scanner’s hard drive and archived. Conventional Method Conventional water fat separation was performed via a multi-point fat-water separation with R2* using a graph cut field map estimation algorithm (1) with the ISMRM water-fat Toolbox (2). Conventional processing of the bipolar acquisition was done separately using the even and odd echo images and results then averaged, producing water, fat, R2* and off-resonance images for each acquisition.
120 images were identified with artifacts and excluded from training. Six imaging sessions, one normal, three chronic and two acute MI patients consisting of 80 acquisitions were also held out for method evaluation and not used in the training process. The images were used for ConvNet prediction and subsequent quantitative analysis of water- and fat-only images, R2* and off-resonance. 1000 acquisitions were then available for training (900 fitting and 100 validation).
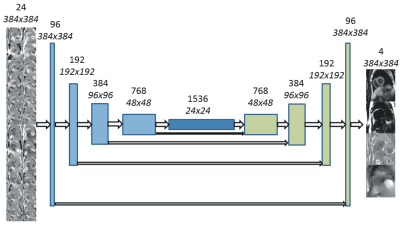
A U-Net (3) convolutional neural network was used for deep learning water-fat separation (Figure 1). Using all available data, the input to the training algorithm was 24 real and imaginary images from 12 echo times used as 24 channels of the ConvNet. Also investigated was the use of magnitude only inputs. Additionally, single echo acquisitions were simulated using only the first opposed-phase echo (TE=2.4 ms). The implementation was realized using Keras 2.0 (30) and TensorFlow 1.2 (31) (both freely available software). An EVGA GEFORCE GTX 1080 Ti graphics card was used for GPU accelerated calculations.
Results
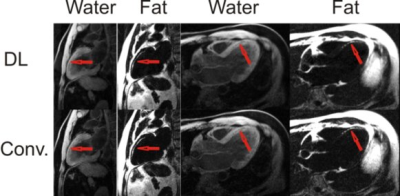
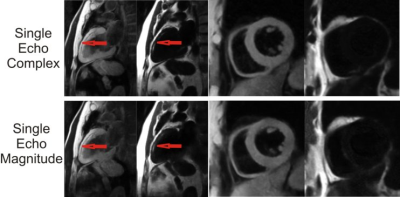
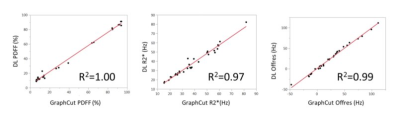
The U-Net ConvNet architecture learned the water-fat separation problem quickly with a small amount of overfitting. Representative examples of DL water-fat prediction over a variety of anatomical slice planes in comparison to the conventional model based method are shown in F. Water-fat separation was visually comparable between deep learning and the conventional model based method. Image resolution was equivalent between methods and well depicted intramyocardial fat deposition (Figure 2) in chronic myocardial infarction. Intramyocardial hemorrhage was well depicted in R2* maps in areas of acute myocardial infarction, while water images showed homogeneous signal (Figure 3). Representative examples of magnitude-only and single-echo separation in the same individuals are shown in Figure 4. Magnitude only performed comparably well to complex separation. Single echo separation did not have the same high quality as 12 echo separation, but performed well as a fat suppression method. There were 26 acquisitions where the conventional model based method failed in inconsistencies of bipolar acquisition. The DL method did not have this problem and performed well without bipolar acquisition artifacts. There was an excellent correlation (R2>=0.97, p<0.001) between the DL and conventional model based method (Figure5) over a wide range of values for PDFF, R2* and off-resonance with 12 complex echoes. Single-echo ConvNets provided good estimations of PDFF, and weak estimates of R2* and off-resonance . The 12 echo magnitude ConvNet provided both excellent estimates of PDFF and R2*, but a weak estimate of off-resonance. Signal-to-noise ratio was consistently higher in DL. In ROI analysis, it was 14% higher (SNR=8.4 vs 9.8), p<0.001.Conclusions
Deep learning utilizing ConvNets is a feasible method for MR water-fat separation imaging with complex, magnitude and single echo image data. A trained U-Net can be efficiently used for MR water-fat separation, providing results comparable to conventional model based methods.Acknowledgements
No acknowledgement found.References
- Hernando D, Liang ZP, Kellman P. Chemical shift-based water/fat separation: a comparison of signal models. Magn Reson Med 2010;64(3):811-822.
- Hu HH, Bornert P, Hernando D, Kellman P, Ma J, Reeder S, Sirlin C. ISMRM workshop on fat-water separation: insights, applications and progress in MRI. Magn Reson Med 2012;68(2):378-388.3. 3.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. MICCAI 2015, p 234-241.
Figures