5519
High SpatiotemporalResolution DCE MRA and Perfusion in a Single 4DAcquisition Exploiting KineticModel Based Signal Priors1Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Republic of Korea, 2Department of Radiology, Gacheon University Gil Medical Center, Incheon, Republic of Korea, 3Department of Radiology, Seoul National University, Seoul, Republic of Korea
Synopsis
In this work we develop a high spatiotemporal resolution (spatial ~ 1.0 mm3, temporal ~ 1.6 sec) simultaneous DCE MRA and perfusion within a single 4D acquisition exploiting kinetic model based signal priors. It is demonstrated that the proposed, high spatiotemporal resolution DCE MRI, which enables rapid sampling of AIF, depicts microvascular permeability in pathological tissues (e.g., tumor) much more accurately than conventional DCE MRI (temporal resolution: 5.0 sec).
Introduction
To comprehensively assess vascular pathologies that may result from either largeor small vessel diseases, it is desirable to investigate both macroscopic arterialand microscopic vascular information. To this end, contrast-enhanced magneticresonance angiography (MRA) and perfusion were typically performed inseparate exams, which requires injection of contrast agents twice and maycorrespondingly increase a contrast dose. Furthermore, low temporal resolution(4 ~ 6 sec) in conventional DCE MRI precludes accurate estimation of arterialinput function (AIF), making it difficult to quantify perfusion/permeability relatedparameters. Given the above considerations, in this work we develop a highspatiotemporal resolution (spatial ~ 1.0 mm 3 , temporal ~ 1.6 sec) simultaneous DCE MRA and perfusion within a single 4D acquisition exploiting kinetic modelbased signal priors. It is demonstrated that the proposed, high spatiotemporalresolution DCE MRI, which enables rapid sampling of AIF, depicts microvascularpermeability in pathological tissues (e.g., tumor) much more accurately thanconventional DCE MRI (temporal resolution: 5.0 sec).Materials and Methods
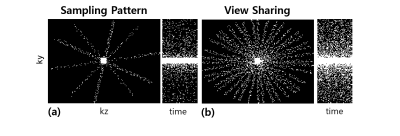
DCE Data Acquisition: Simultaneous acquisition of DCE MRA and perfusion data were performed in patients on a 3.0 T whole body MR scanner (Skyra, Siemens Healthcare) using multi-phase spoiled 3D GRE with high spatiotemporal resolution (spatial ~ 1.0mm3, temporal ~ 1.6 sec). Data were vastly under-sampled in a radial-like pattern on cartesian grid (R = 50) and then shared in the peripheral k-space over two or three neighboring phases for reconstruction. The imaging parameters were: TR/TE = 3.05/1.3ms, flip angle=15◦, matrix size = 92×192×144, spatial resolution = 1.1×1.1×1.1mm3, temporal resolution= 1.65sec, number of phases = 180, bandwidth/pixel = 650Hz/pixel, asymmteric Echo = 70% in the readout direction. For accurate quantitative analysis of perfusion, B1 maps and reference T10 maps were acquired using AFI (TRs =7ms, 35ms) [1] and variable-flip-angle imaging (3°,15°) [2], respectively.
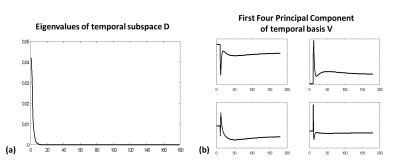
DCE Signal Model: Concentration time-course in DCE MRI can be described by using either Patlak [3] or extended Tofts-Ketty model [3]. Given the concentration time-course, temporal signal profiles in DCE MRI are then simulated, row-vectorized, and stacked into a signal library: $$$D=UΣV^H$$$ where $$$D$$$ consists of simulated temporal signal profile vectors, $$$U$$$ is the basis of the column vectors in $$$D$$$, $$$V$$$ is the basis of the row vectors in $$$D$$$ and is used as the temporal basis for the signal time-course, and $$$Σ$$$ is the singular value matrix. Figure 1 represents the variation of eigenvalues in and its corresponding eigenvectors in the temporal basis . Since most of singular values are close to zero, the concentration time-course can be synthesized using only a few principal eigenvectors in $$$V$$$. Given the discrepancy of modeled signals and measured signals, the proposed DCE signal model can be written by: $$X=X_0 + X_D + X_M + N $$ where $$$X$$$ is the Casorati matrix that an image column vector in each time frame is stacked column-wise over the entire dynamic phases, $$$X_0$$$ is the reference matrix consisting of the mean column vector by averaging the pre-contrast images, $$$X_D$$$ is the signal of interest, and $$$X_M$$$ is the leakages of signals (residual artifacts).
DCE Reconstruction with Kinetic Model Based Signal Priors: The proposed DCE reconstruction from incomplete measurements is performed by solving the following constrained optimization problem with kinetic model based signal priors:
$$(\hat{X_{D}},\hat{U},\hat{X_{M}})= \arg \underset{X_{D},U,X_{M}}{min}\left \| D_{t}X_{D} \right \|_{1}+\lambda_{U}\left \|D_{s}U \right \|_{1}+\lambda _{M}\left \| \Psi X_{M} \right \|_{1} $$$$ s.t. d_{r}=E(X_{D}+X_{M}), X_{D}=UV_{r} $$
Results
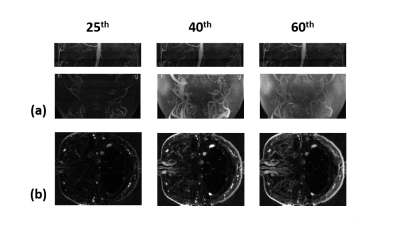
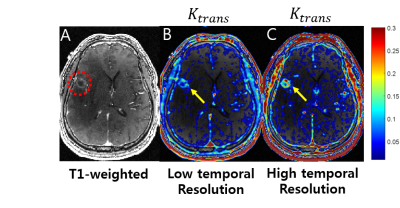
Fig. 2 shows the sampling pattern in k-t space. View-sharing was performed in the corresponding space. Fig. 3 shows MIP images obtained using the proposed reconstruction algorithm with R=50. Fig. 4 shows perfusion parameters derived permeability parameters for discriminating brain tumor between different temporal resolution. Compared with the low temporal resolution, high temporal resolution shows superior result of Ktrans .The proposed method can lead to capture both macroscopic arterial and microscopic vascular information simultaneously.
Conclusion
We successfully demonstrated that the proposed method enables high spatiotemporal resolution (1.0 mm3, 1.6 sec), simultaneous DCE MRA and perfusion within a single 4D acquisition, leading to rapid sampling of AIF and thus accurate quantification of permeability information in pathological tissues. It is expected that the proposed method will widen its clinical utilities due to accurate, simultaneous depiction of large and small vascular information and thereby reduction of injected contrast dose.Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF)funded by the Ministry of Science (2016M3C7A1913844, NRF-2017R1A2B4012581).References
1.Vasily L, Yarnykh. Actual Flip-Angle Imaging in the Pulsed steady state : A Method for Rapid Three-Dimensional Mapping of the Transmitted Radiofrequency
2.FieldSean C.L Deoni, PhD :High-Resolution T1 Mapping of the Brain at 3T with Driven Equilibrium Single Pulse Observation of T1 with High speed Incorporation of RF Field Inhomogeneities
3.Anna K. Heye, Ross D. Culling, Maria del C. Valdés Hernández, Michael J. Thrippleton, Joanna M.Wardlaw : Assessment of blood-brain barrier disruption using dynamic contrast-enhanced MRI. A systematic review
4.Marios Spanakis , Eleftherios Kontopodis , Sophie Van Cauter , Vangelis Sakkalis , Kostas Marias : Assessment of DCE–MRI parameters for brain tumors through implementation of physiologically–based pharmacokinetic model approaches for Gd-DOTA
Figures