5510
Self-referenced DCE-MRI: reference region modelling without a reference tissue1Medical Physics Unit, McGill University, Montreal, QC, Canada, 2Research Institute of the McGill University Health Centre, Montreal, QC, Canada
Synopsis
Analysis of dynamic contrast enhanced (DCE) MRI using the reference region model (RRM) requires manual identification of a reference tissue, which is typically chosen as muscle external to the tissue of interest. The current work proposes an automated technique, named the self-reference approach, for finding a suitable reference-region curve from within the tissue of interest. This approach was evaluated on in-vivo data from glioblastoma and sarcoma patients, and was found to be equally as effective as manual identification of the reference tissue in most cases, and in some cases provides substantially improved fits.
Introduction
Analysis of dynamic contrast-enhanced (DCE) MRI, notably using the reference region model (RRM), provides quantitative information about blood supply in tissue. The RRM does not require an arterial input function (AIF), an advantage over other models such as the conventional Tofts model. Instead of an AIF, the RRM uses the tracer concentration in a selected reference region – typically muscle – with the assumption that the reference region and the tissue of interest share the same AIF.1
A first limitation of the RRM is the need to manually identify a reference tissue. Sometimes, no suitable reference tissue is in the imaging field of view. Manual selection can also introduce variability as different operators might identify different reference regions. A second limitation is that the selected reference region might not share the same AIF as the tissue of interest.
The current work addressed these two limitations by proposing an automated technique for identifying a suitable reference tissue curve from within the tissue of interest. This is named the self-reference approach. This novel method is demonstrated on in-vivo DCE-MRI data from glioblastoma multiforme and soft tissue sarcoma patients.
Theory
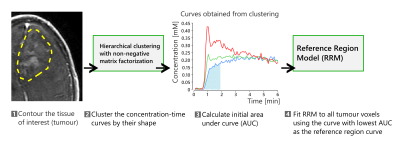
The self-reference approach uses a hierarchical clustering algorithm that relies on non-negative matrix factorization.2 This algorithm clusters the concentration-time curves by their shape into N clusters without taking spatial information into account. N is user-defined, and the current work uses N=3.
Figure 1 shows a summary of the steps in the self-reference approach. First, the tissue of interest is contoured. The concentration-time curves in the tissue of interest are then fed to the clustering algorithm, which provides N=3 concentration-time curves that are potential candidates for the time-course in the reference region. The RRM requires that the reference region be weakly vascularized, so the candidate with the lowest initial area under the curve is chosen and then used with the RRM.
Methods
The proposed self-reference region approach was evaluated on two sets of in-vivo DCE-MRI data.
The first dataset is from three patients with glioblastoma multiforme.3 This data was acquired at 3 T (3D SPGR sequence, temporal resolution ~5 seconds, duration of 6 minutes). A second dataset was selected from a local study on soft tissue sarcoma. This data was acquired at 1.5 T (3D SPGR sequence, temporal resolution ~10 seconds, duration of 9 minutes).
Two versions of the RRM were fitted to the data, one using manually identified muscle as the reference region, and the other using the self-reference approach. For comparison, the extended Tofts model was also fitted. The AIF for the Tofts model was measured for fits to glioblastoma data (assumed hematocrit = 0.45), while fits to sarcoma data used a model-based AIF.4 The steady-state part of the AIF was also used to scale the maps from the RRM.5
Results and Discussion
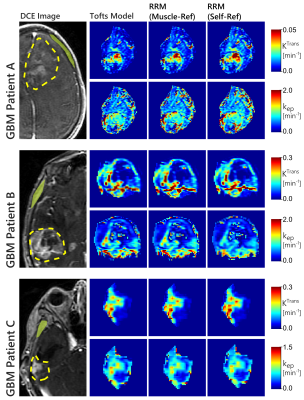
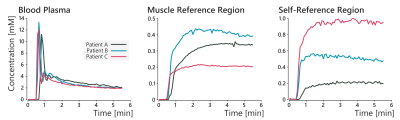
In the glioblastoma dataset, both the manual and self-reference approaches produced similar maps that were also in agreement with the Tofts model, as seen in figure 2. The concentration-time curves for the AIF and reference region curves for these patients is shown in figure 3. These results show that the self-reference approach works just as well as a manually identified reference region.
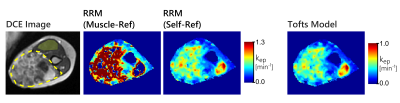
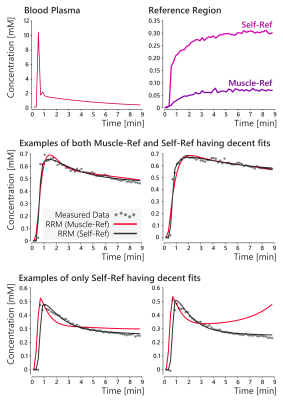
In the sarcoma data however, the two RRM approaches did not always agree with one another. An example of this disagreement is shown in figure 4, where only the automated self-reference approach provided maps that agreed well with the Tofts model. Concentration-time curves for the AIF and reference region for this patient are shown in figure 5. This figure also shows examples of the RRM fits, with two cases where both RRM versions have decent fits, and two cases where only the proposed self-reference approach provided decent fits. The average norm of the residuals in the tumour was 0.28 with muscle as the reference tissue, and 0.09 with the self-reference approach.
Conclusion
Self-referenced DCE-MRI identifies a reference region curve from within the tissue of interest to be used with the reference region model. This simplifies the analysis as the manual identification of a reference tissue is not required. This approach performs as well or better than the conventional approach of manual identification of a reference tissue.
Acknowledgements
Funding for this work was provided by the RI-MUHC (Montreal General Hospital Foundation), an NSERC Discovery Grant, and the NSERC CREATE Medical Physics Regional Training Network (Grant no. 432290). Sarcoma data collected with the help of colleagues at the MUHC (PI: Dr. C. Freeman). The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.References
1. Yankeelov et al. MRI 2005; 23:519–29. doi: 10.1016/j.mri.2005.02.013.
2. Gillis, Kuang, & Park (2015). IEEE Transactions on Geoscience and Remote Sensing, 53(4), 2066–2078. http://doi.org/10.1109/TGRS.2014.2352857
3. Scarpace, Mikkelsen, Cha, et al. Radiology Data from The Cancer Genome Atlas Glioblastoma Multiforme [TCGA-GBM] Collection. 2016. doi: 10.7937/K9/TCIA.2016.RNYFUYE9.
4. Parker et al. MRM 2006;56:993–1000. doi: 10.1002/mrm.21066.
5. Ahmed & Levesque, Proc. Intl. Soc. Mag. Reson. Med. 25 (2017), 1907
Figures