5450
Block-Interleaved Segmented EPI for voxel-wise high-resolution fMRI studies at 7T1CiNet, NICT, Suita-shi,Osaka, Japan
Synopsis
We report a novel segmented EPI method preserving the pattern of hemodynamic response in bock design fMRI studies. The proposed method uses an EPI-based anatomical data obtained with the same distortion from task EPI scans for further fMRI analysis. At sub-millimeter spatial resolution, the proposed method avoids functional to structural registration and spatial smoothing giving us an opportunity to investigate activity dynamics at the columnar level spatial resolution with high specificity. Our results showed that the proposed method can perform voxel-wise high-resolution fMRI studies with a voxel size of 0.6×0.6×0.6 mm3 at 7T.
Purpose
Ultrahigh-field (≥7T) high-resolution functional MRI (fMRI) has shown its importance for studying the function of subcortical structures in the brain. However, voxel-wise high-resolution fMRI is still challenging because of several reasons: the reduced temporal signal-to-ratio (tSNR) due to prominant physiological noise, the low temporal resolution, and the geometric dissimilarities of the functional compared to the anatomical data. We presented a method to reduce physiological noise at the last ISMRM meeting [1]. In this work, we report a novel segmented EPI method called Block-Interleaved Segmented EPI (BISEPI) that allows us to investigate sub-millimeter-level BOLD dynamics acquired within reasonably-acceptable scan time. Further, the proposed method can also acquire structural images that do not require any pre-processing such as distortion correction for registration. At sub-millimeter spatial resolution, the proposed method avoids functional to structural registration and spatial smoothing giving us an opportunity to investigate activity dynamics at the columnar level spatial resolution with high specificity. Our results showed that the proposed method can perform voxel-wise high-resolution fMRI studies with a voxel size of 0.6×0.6×0.6 mm3 at 7T.Methods
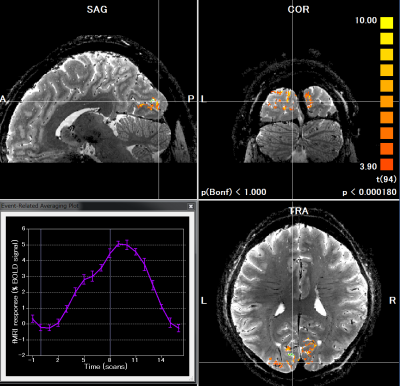
The low temporal resolution in conventional segmented EPI is not suitable for detecting BOLD activities, because the detailed pattern of hemodynamic response is masked by the time it takes to acquire one image (e.g. it would take 20 seconds for 20 shot EPI to obtain one image). Here, we propose a novel method that acquires all shot #1 data for one ON/OFF set (8/8 volumes in this experiment) first then shot #2, #3…#20. In post-processing, all data from shot #1 to shot #20 of one volume (same timing-point in block design) are reconstructed to images of one volume. A human brain was scanned using BISEPI sequence on a Siemens MAGNETOM 7T scanner with a 32-channel phased array head coil (Nova Medical, MA, USA) to obtain 1920 segmented EPI data (96 volumes, segment 20, scan time ~ 32 min) at spatial resolution of 0.6×0.6×0.6 mm3 followed by two structural scans EPI with the same spatial resolution. Acquisition parameters for functional/structural scans were as follows: TR = 1000/6000 ms, TE = 21 ms, flip angle (FA) = 52°/90°, slice = 22/128, averages = 1/3. A standard block design task of checkerboard visual stimulation was performed in the experiment with stimulation ON and OFF for blocks of 8 s each, total 16 shots for one ON/OFF unit and repeated 120 times. In other words, there are 16 timing points for detecting the BOLD response in one ON/OFF unit. Two structural EPI scans (128 slices each) were acquired to cover the whole brain using the same sequence and same parameters except TR, FA and averages. fMRI data were reconstructed from raw data in Matlab prior to analyzing in BrainVoyager QX (Brain Innovation, Maastricht, The Netherlands). The following pre-statistics processing was applied; interscan slice time correction, high-pass filtering using a general linear model (GLM) Fourier bases set of 2 sine/cosines as well as temporal Gaussian smoothing with a full width half maximum (FWHM) kernel of 2 data points. No spatial smoothing was performed. Generalized linear model (GLM) analysis was applied with stimulation ON/OFF as a binary regression variable.
Results and Discussion
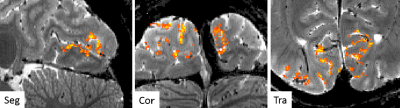
Figure 1 shows the task-related activations obtained by the proposed method; functional 20-shot EPI overlaid on the structural 20-shot EPI. Actual hemodynamic response observed at certain activated voxels is also plotted. Unlike conventional segmented EPI approach, the novel segmented EPI can observe hemodynamic response within reasonably acceptable scan time. Figure 2 highlights activated voxels in the visual cortex in detail. One of the strength of the proposed method is that functional EPI and structural EPI are both acquired by the same pulse sequence and imaging parameters except only for flip angle and TR, both of which are geometry-independent. Therefore, there is no need to perform distortion correction and even registration. Moreover, spatial smoothing is not necessary because our segmented EPI provides a high tSNR, allowing us to better investigate voxel-wise dynamics at cortical layers. These results demonstrate the feasibility of the proposed method for conducting sub-millimeter voxel-wise high-resolution fMRI studies. The proposed method can easily be expected to transform EPI structural data into a T1-like image by segmentation tools and inverting the gray-white matter contrast and to generate and inflate cortical surfaces.