5445
Human Connectome Project (HCP)-style resting state functional MRI at 7 Tesla using RF parallel transmission1Medical School, University of Minnesota, Minneapolis, MN, United States
Synopsis
A major component of the Human Connectome Project (HCP) in the WU-Minn consortium is slice-accelerated whole-brain resting-state functional MRI (rfMRI) at both 3T and 7T. Although providing better contrast and higher spatial resolution, the 7T acquisition is compromised by RF nonuniformity. Here, we demonstrate how RF parallel transmission (pTx) can be used to improve RF uniformity for highly-accelerated, HCP-style rfMRI at 7T with simultaneous multislice multiband approach. Our results demonstrate that the combination of pTx with the 8-channel transmit/32-channel receive coil (Nova8Tx/32Rx) enhanced functional contrast-to-noise ratio (fCNR) in most cortical surfaces and subcortical voxels relative to the original HCP protocol utilizing the Nova1Tx/32Rx in combination with dielectric padding. The enhanced fCNR in turn yielded higher correlation values in seed-based connectivity measures. These results demonstrate that pTx provides significant gains in whole-brain rfMRI employed for defining the functional connectome of the human brain.
Purpose
To investigate how RF parallel transmission (pTx) can be employed to improve Human Connectome Project (HCP)-style, highly-accelerated, whole-brain, resting-state functional MRI (rfMRI) at 7 Tesla (7T), and provide a comparison to the 7T HCP-style data acquisition using a single-channel transmit coil and dielectric padding.Methods
We conducted human experiments on a 7T MR scanner (Siemens, Erlangen, Germany), which can be operated in a single-transmit (1Tx) or pTx mode, both with 32-channel receive capability.
Three healthy subjects who signed a consent form approved by the local Institutional Review Board were scanned. Each subject was scanned twice: once in the pTx mode using the Nova 8Tx/32Rx head coil and once in the 1Tx mode using the Nova 1Tx/32Rx head coil plus dielectric padding as employed in the original HCP 7T protocol.
For pTx acquisition, the Nova 8Tx/32Rx coil was used in the “protected” mode to ensure RF safety. Additionally, band-specific pTx multiband (MB) RF pulses with single spokes were designed using the slab-wise design framework (1), optimizing for flip angle homogeneity and minimizing local SAR.
To capitalize on the coil geometry and promote RF performance, pulses were designed to image sagittal slices (2) as opposed to the axial oblique slices employed in the original HCP. Based on the MB sequence used in the HCP, a new pTx-enabled, 2D echo-planar-imaging (EPI) sequence suitable for BOLD fMRI acquisitions was developed to enable application of multiple pTx MB RF pulses for excitation.
For each subject, we acquired full HCP-style rfMRI data using the original HCP 7T acquisition parameters and methods (defined here as HCP protocol) (3); additionally, data were acquired employing the same protocol except for the use of pTx pulses and the use of sagittal slices (defined here as pTx-HCP protocol).
For the HCP protocol, data were obtained in four runs, each containing a timeseries of 900 images (a total of 3600 images). Each run was acquired with 1.6-mm isotropic resolution, MB5 (i.e. 5-fold slice acceleration), 2-fold acceleration along the phase encode direction (i.e. iPAT2), 7/8 partial Fourier, inter-slice shift=FOV/3, bandwidth=1924 Hz/pixel, TR/TE=1000/22 ms. To reduce the susceptibility-induced signal dropout in the final concatenated time series (4), the four runs were acquired with inverted phase encode directions and the total acquisition time was ~65 minutes. For correction of EPI distortions and bias fields, spin-echo EPI datasets with same resolution and echo spacing were acquired.
Like the HCP protocol, our pTx-HCP protocol also acquired four runs of rfMRI data (900 images each) from each subject. The use of sagittal slices required more slices (90 vs. 85) to be acquired in the left-right dimension to ensure whole-brain coverage. To keep the same TR of 1 second, pulse length was reduced to 5440 µs (as opposed to 5760 us for the HCP protocol).
During data acquisition, subjects were instructed to fixate at a black cross displayed over a white background.
All rfMRI data underwent the HCP spatial (5) and temporal (4) preprocessing pipelines. As required by the HCP preprocessing pipelines, high-resolution whole-brain T1-weighted (T1w) and T2-weighted (T2w) structural images were acquired, both with 0.7-mm isotropic resolution and using the Nova 1Tx/32Rx coil and dielectric padding.
Results
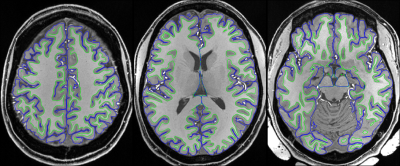
Despite of the RF nonuniformity across the brain, satisfactory definitions of pial and white surfaces were achieved in most cortical regions when using Nova 1Tx/32Rx and dielectric padding for the T1w and T2w structural acquisitions (Fig. 1).
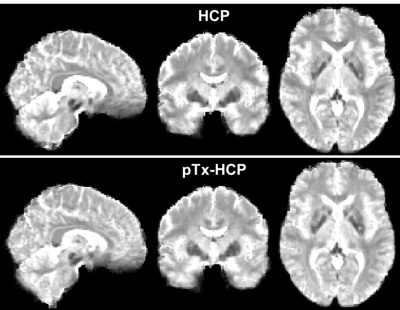
After undergoing the HCP preprocessing pipelines, the rfMRI data presented little residual distortion or signal dropout (Fig. 2).
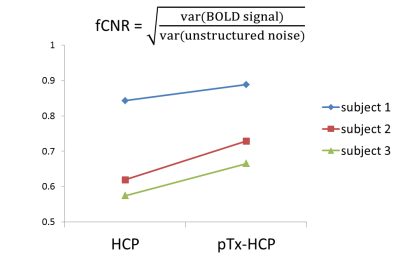
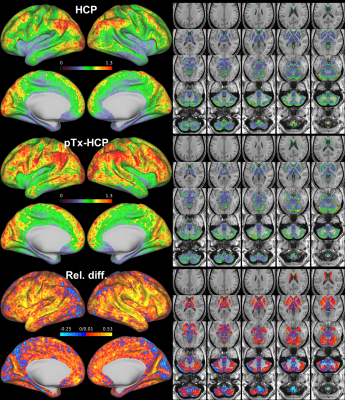
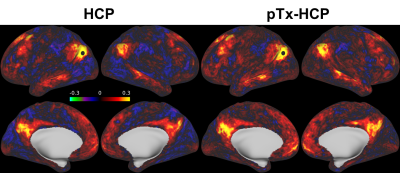
Further data analysis revealed that the use of pTx enhanced fCNR in all three subjects (Fig. 3). Spatially, such enhancement of fCNR was observed in most cortical surfaces and subcortical voxels (Fig. 4).
Further, the enhancement of fCNR translated into increased correlation values across the cortical surfaces when placing a seed in the posterior parietal cortex (known to be a part of the default mode networks) to estimate the seed based dense connectome (Fig. 5).
Discussion
Our results show that pTx can be used to improve the functional contrast for 7T HCP-style rfMRI data and functional connectivity measures. Future work will examine how pTx can be used to further improve flip angle uniformity by using multispoke pTx MB pulses and further accelerate rfMRI by allowing higher MB factors (because of SAR reduction).Conclusion
We have demonstrated the advantage of pTx over single-transmit for HCP-style rfMRI at 7T. Our data demonstrate that pTx can enhance fCNR in most cortical surfaces and subcortical voxels and improve functional connectivity measures compared to the original HCP 7T protocol. These data demonstrate that pTx provides significant advantages for HCP-style whole-brain rfMRI data that are desired in many neuroscience and clinical applications.Acknowledgements
This work was supported by NIH grants including P41 EB015894, U01 EB025144 and P30 NS076408.References
1. Wu X, Schmitter S, Auerbach EJ, Ugurbil K, Van de Moortele PF. A generalized slab-wise framework for parallel transmit multiband RF pulse design. Magn Reson Med 2016;75(4):1444-1456.
2. Wu X, Tian J, Schmitter S, Vaughan JT, Ugurbil K, Van de Moortele PF. Distributing coil elements in three dimensions enhances parallel transmission multiband RF performance: A simulation study in the human brain at 7 Tesla. Magn Reson Med 2016;75(6):2464-2472.
3. T. Vu A, Jamison K, Glasser MF, Smith SM, Coalson T, Moeller S, Auerbach EJ, Uğurbil K, Yacoub E. Tradeoffs in pushing the spatial resolution of fMRI for the 7T Human Connectome Project. NeuroImage 2017;154(Supplement C):23-32.
4. Smith SM, Beckmann CF, Andersson J, Auerbach EJ, Bijsterbosch J, Douaud G, Duff E, Feinberg DA, Griffanti L, Harms MP, Kelly M, Laumann T, Miller KL, Moeller S, Petersen S, Power J, Salimi-Khorshidi G, Snyder AZ, Vu AT, Woolrich MW, Xu J, Yacoub E, Uğurbil K, Van Essen DC, Glasser MF. Resting-state fMRI in the Human Connectome Project. NeuroImage 2013;80(0):144-168.
5. Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 2013;80(0):105-124.
Figures