5429
Automated Extraction of the Fetal Brain from Functional MRI Data1Psychiatry, University of Michigan, Ann Arbor, MI, United States, 2Wayne State University, Detroit, MI, United States
Synopsis
In this study, we present a novel application of a Convolution Neural Network algorithm to a challenging image segmentation problem: fetal brain segmentation. Resting-state fMRI data was obtained from 192 fetuses (gestational age 20-40 weeks, M=31.9, SD=4.28). The output from automated extractions are compared with the ground truth of manually drawn brain masks. We report that automated fetal brain localization and extraction is achievable at the same integrity of manual methods, in a fraction of the time.
Introduction
Recent advances in resting-state functional magnetic resonance imaging (rs-fMRI) have enabled observation of the human brain in utero, a period of development previously inaccessible. This provides a window into the nature and manner in which the human brain’s architecture is initially assembled [1,2]. Many tools and software packages exist for the processing and analyzing of rs-fMRI data. However, the recent application of fMRI to studying the fetal brain has produced some unique challenges that traditional preprocessing methods cannot currently address. Critical issues include: high levels of motion, multiple head orientations across a time series, extraction of fetal brain tissue from surrounding maternal tissue, and reorientation of all volumes to a standard space. In this study, we adapt a Convolutional Neural Network (CNN) algorithm for automated fetal brain identification and extraction, and we evaluate its performance. CNN algorithms are powerful learning routines that can identify complex, highly non-linear patterns in spatially structured high dimensional datasets, and they are increasingly utilized in processing applications in both medical and non-medical settings [3,4].Methods and Results
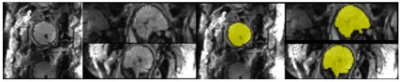
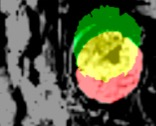
In the application of fetal brain extraction, prior work has used similar techniques on structural T2-weighted images [5,6,7]. Here we demonstrate, for the first time, that these methods are applicable to brain extraction from rs-fMRI volumes (EPI BOLD TR/TE, 2000/30 ms; 2 runs, 6min; 4mm slice thickness; axial; SAR = 0.3). Fetal MR exams were performed with a Siemens Verio 3T scanner using an abdominal 4-Channel Flex Coil. Resting-state data was obtained from 192 fetuses (gestational age 20-40 weeks, M=31.9, SD=4.28). From each subject, periods of low fetal movement were identified (at least 10 TRs in length) and a reference volume to be masked was chosen. This process resulted in 1,210 volumes and their corresponding hand drawn masks (Fig 1). To determine accuracy, the output from automated extractions are compared with the ground truth of manually drawn brain masks. The results indicate that our CNN algorithm generates fetal brain masks of similar quality to manual methods in significantly less time. Furthermore, we show that our methods can identify the brain in every volume of a time-series, and can be used to calculate frame-wise displacement, an important variable for resting-state connectivity analysis (Fig 2). These motion parameters are also a valuable asset for subsequent brain reorientation to standard space. Issues associated with training the CNN and artifact management will be discussed.Discussion
Fetal fMRI is an emerging field of research in need of specialized tools. The current standard for fetal fMRI data processing is time consuming and open to human error. Large scale projects, such as the Developing Human Connectome Project, have collected more than 600 fetal fMRI scans. Manual preprocessing of such data is not practical, as a single hand drawn mask can take up to 4 hours. An automated preprocessing pipeline can mitigate bias, reduce errors, and facilitate inter-study comparisons of rs-fMRI findings. All code from this study is publicly available on GitHub [8]. Future work will expand our automated pipeline to include reorientation to standard template space.Conclusion
Our adaptation of the CNN algorithm to localize and extract the fetal brain directly from each volume of a functional time series is a crucial first step in for a future comprehensive automated preprocessing pipeline for fetal imaging. Our methods have built upon and improved existing pipelines to significantly reduce the time cost, while matching the integrity of gold standard manual methods. Application of this method will potentially facilitate observations of early brain development and aid research in identifying the origins of common developmental disorders, such as autism and ADHD.Acknowledgements
We thank the research assistants who contributed many hours to hand drawing brain masks. The authors also thank participant families who generously shared their time.References
[1] Jakab, A., Schwartz, E., Kasprian, G., Gruber, G. M., Prayer, D., Schöpf, V., & Langs, G. (2014). Fetal functional imaging portrays heterogeneous development of emerging human brain networks. Frontiers in Human Neuroscience, 8, 852. http://doi.org/10.3389/fnhum.2014.00852
[2] Thomason, M. E., Grove, L. E., Lozon, T. A., Vila, A. M., Ye, Y., Nye, M. J., Romero, R. (2015). Age-Related Increases in Long-Range Connectivity in Fetal Functional Neural Connectivity Networks In Utero. Developmental Cognitive Neuroscience, 11, 96–104. http://doi.org/10.1016/j.dcn.2014.09.001
[3] Zeiler, M. D., & Fergus, R. (2014). Visualizing and Understanding Convolutional Networks. In D. Fleet, T. Pajdla, B. Schiele, & T. Tuytelaars (Eds.), Computer Vision – ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part I (pp. 818–833). Cham: Springer International Publishing. https://doi.org/10.1007/978-3-319-10590-1_53
[4] M. Egmont-Petersen, D. de Ridder, H. Handels, Image processing with neural networks—a review, In Pattern Recognition, Volume 35, Issue 10, 2002, Pages 2279-2301, ISSN 0031-3203, https://doi.org/10.1016/S0031-3203(01)00178-9.
[5] Salehi, S. S. M., Erdogmus, D., & Gholipour, A. (2017). Auto-context Convolutional Neural Network (Auto-Net) for Brain Extraction in Magnetic Resonance Imaging. IEEE Transactions on Medical Imaging, PP(99), 1–1. https://doi.org/10.1109/TMI.2017.2721362
[6] Ison, M., Donner, R., Dittrich, E., Kasprian, G., Prayer, D., & Langs, G. (2012). Fully automated brain extraction and orientation in raw fetal MRI. In Workshop on Paediatric and Perinatal Imaging, MICCAI (pp. 17–24).
[7] Tourbier, S., Velasco-Annis, C., Taimouri, V., Hagmann, P., Meuli, R., Warfield, S. K., … Gholipour, A. (2017). Automated template-based brain localization and extraction for fetal brain MRI reconstruction. NeuroImage, 155, 460–472. https://doi.org/10.1016/j.neuroimage.2017.04.004
[8] https://github.com/saigerutherford/fetal-code
Figures