5391
Semi-automatic, Machine Learning Segmentation of Peripheral Nerves in Healthy Volunteers and Patients1Institute for Surgical Technology and Biomechanics, University of Bern, Bern, Switzerland, 2Support Center for Advanced Neuroimaging (SCAN), Institute for Diagnostic and Interventional Neuroradiology, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland, 3Department of Neurology, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland
Synopsis
Magnetic resonance neurography (MRN) is increasingly used to diagnose peripheral neuropathy. Here, we propose a semi-automatic multimodal machine learning-based segmentation algorithm to segment peripheral nerves from MRN images. Our algorithm was tested on 9 volunteers and 25 patient cases suffering from sciatic neuropathy. Compared to manual segmentation, Dice coefficients were 0.723 ± 0.202 and 0.443 ± 0.228, respectively, with segmentation times of 5 ± 1 for semi-automatic, and 24 ± 8 minutes for manual segmentation. Our preliminary results suggest that machine learning-based segmentation of the sciatic nerve is possible in healthy and diseased nerves in clinically feasible time.
Purpose
Magnetic
resonance neurography (MRN) has emerged as a complementary diagnostic tool for
peripheral neuropathy to the state-of-the-art relying on neurological
examination and electrodiagnostic studies (EDS). MRN provides advantages
compared to EDS such that it enables physicians to assess deeply situated
nerves and nerves close to the trunk including their surrounding tissues.
However, MRN remains a qualitative approach despite that a quantification could
assist in the diagnosis and monitoring of peripheral neuropathies in terms of
biomarkers calculated from segmented peripheral nerves1. Therefore,
we developed a semi-automated, yet time-effective and hence clinical feasible
segmentation approach based on machine learning to segment peripheral nerves
from MRN images of extremitiesMethods
Data: The upper leg of 9 healthy volunteers (3 female, 6 male, age = 24 ± 3) and 25 patients (15 females, 10 males, age = 54 ± 15) suffering from clinically diagnosed sciatic neuropathy were imaged.
MR methods: A standard turbo inversion recovery magnitude (TIRM) and spin echo T2-weighted (T2w) sequence acquired with a circular 15-channel knee coil in a MR scanner running at 3T (MAGNETOM Verio, Siemens Healthcare GmbH, Erlangen, Germany) were used for axial imaging, with following parameters: TIRM (TR = 4500 ms, TE = 32 ms, TI = 210 ms, FOV = 256×256 mm2, voxel size = 0.78×0.78×4.0 mm3, 60 slices); T2w (TR = 4690 ms, TE = 82 ms, FOV = 384×330 mm2, voxel size = 0.52×0.52×4.0 mm3, 60 slices).
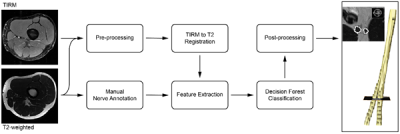
Segmentation method: We propose a semi-automatic, machine learning-based, multi-sequence pipeline (Figure 1), using as input the two sequences and seeds in the first and last slice of the T2w stack provided by the user identifying the proximal and distal ends of the nerve. The output is a binary segmentation that identifies the peripheral nerve and background. The pipeline consists of five main steps: (i) pre-processing of the images using bias field correction2 and normalization to zero mean and unit variance, (ii) registration of the TIRM to the T2w, (iii) extraction of context- and intensity-based features, (iv) voxel-wise tissue classification using a decision forest3, and (v) regularization using a dense conditional random field4.
Result analysis: We evaluated the performance of our method in both cohorts independently using a leave-one-out cross-validation and calculated the Dice coefficient as evaluation metric, using as ground truth manually segmented nerves by an expert. Furthermore, we report the time taken for ground truth generation and calculation using our method.
Results
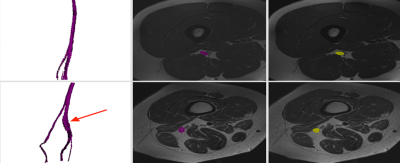
Our semi-automatic segmentation pipeline allowed segmentation of both healthy and diseased nerves. Our segmentation pipeline showed higher results in healthy volunteers than in patients, with Dice coefficients of 0.723 ± 0.202 and 0.443 ± 0.228, respectively. Additionally, the shape of the segmented sciatic nerves is qualitatively consistent with the manually segmented ground truth, and allows depiction of nerve lesions when nerve diameter changes were present (Figure 2). The execution time of the full pipeline takes 5 ± 1 minutes including manual seed annotation and 24 ± 8 minutes for manual segmentation.Discussion
Our semi-automatic, machine learning-based segmentation of the sciatic nerve in the upper leg is possible in both healthy volunteers and patients suffering from neuropathy. Furthermore, the speed increase of the semi-automatic segmentation pipeline by 5-fold makes this approach feasible for clinical use. The drop in performance on the patient cohort is due to common segmentation issues such as over- and under-segmentation, which are present in regions where the nerves do not exhibit a distinctly visible boundary and where patient movement was noticeable. Furthermore, this effect is increased where the nerve has volume fractions smaller than 0.15 % and partially low contrast between the nerve and its surrounding tissues.Conclusion
Semi-automatic segmentation of peripheral nerves is a promising tool and first step towards quantitative evaluation in healthy volunteers and patients suffering from peripheral neuropathies. Future work will focus on the detection of lesions to extract imaging biomarkers and the improvement of the segmentation performance by increasing the number of subjects and the study of more rich features for the training method.Acknowledgements
This research was supported by the Swiss Foundation for Research on Muscle Diseases (ssem).References
1Felisaz PF, Maugeri G, Busi V, et al. MR Micro-Neurography and a Segmentation Protocol Applied to Diabetic Neuropathy. Radiol Res Pract. 2017;2017:1-7.
2Tustison NJ, Avants BB, Cook PA, et al. N4ITK: Improved N3 Bias Correction. IEEE Trans Med Imaging. 2010;29(6):1310-1320.
3Breiman L. Random Forests. Mach Learn. 2001;45(1):5-32.
4Krähenbühl P, Koltun V. Efficient Inference in Fully Connected CRFs with Gaussian Edge Potentials. In: Shawe-Taylor J, Zemel RS, Bartlett PL, Pereira F, Weinberger KQ, eds. Advances in Neural Information Processing Systems 24. Curran Associates, Inc.; 2011:109-117.
Figures