5372
Using Double Diffusion Encoding (DDE) MRI to study tissue microstructure in traumatic brain injury (TBI)1Section on Quantitative Imaging and Tissue Sciences, NICHD, National Institutes of Health, Bethesda, MD, United States, 2Center for Neuroscience and Regenerative Medicine, Uniform Service University of the Health Sciences, Bethesda, MD, United States, 3Quantitative Medical Imagion Section, NIBIB, National Institutes of Health, Bethesda, MD, United States, 4Department of Neurology, University of Pennsylvania, Philadelphia, PA, United States, 5Microscopy and Imaging Core, NICHD, National Institutes of Health, Bethesda, MD, United States, 6Laboratory of Molecular Signaling, NIAAA, National Institutes of Health, Rockville, MD, United States
Synopsis
A double diffusion encoding (DDE) MRI method was used to estimate apparent mean axon diameter (AMD) in a mild traumatic brain injury (mTBI) mouse model exhibitig diffuse axonal injury (DAI). MRI data show clear tissue alterations caused by the injury, while immunohistochemistry data confirm the MRI findings. DDE could potentially be used as a non-invasive means to detect mTBI.
Introduction
Diffuse axonal injury (DAI) is a common pathology present in about 40%-50% of traumatic brain injuries (TBI). DAI1 ranges from mild to severe, causing microstructural changes (some on the scale of microns) within the brain, including cell varicosities, microglia cell migration, and tissue loss2,3. While clinical MRI is routinely used to detect robust physiological and vascular abnormalities, the subtle white matter changes associated with this injury are challenging to diagnose and characterize using conventional MRI methods. Often DAI can only be detected at autopsy1. Diffusion MRI methods show promise to probe subtle changes in brain microstructure4,5. Diffusion correlation methods such as double diffusion encoding (DDE)6,7, in which the signal is sensitized to net displacement correlations rather than the mean-squared net displacement, may improve both sensitivity and specificity in detecting brain abnormalities following injury. We present the use of DDE method to estimate the apparent mean axon diameter (AMD) in a closed-head impact mouse model of engineered rotation acceleration (CHIMERA)3.Materials and Methods
Repeat CHIMERA injury3 was performed three times in a twenty-four hour interval. Two levels of force were applied to the vertex of the head: 0J (sham), and 0.5J. Mice were sacrificed one week after the third injury and perfuse fixed with 4% formaldehyde in PBS. Brains were rehydrated in PBS prior to scanning. DDE filtered MRI data were acquired using a 3D EPI MRI sequence with the following MRI parameters: TE/TR=23/700 ms, 8 segments, 4 averages, voxel resolution =100mm3; DDE NMR parameters: τm = 0, δ = 3 ms, Δ = 20 ms. Gradient orientations were uniformly distributed on a hemisphere; φ the angle between the two pulsed-field gradient (PFG) blocks was applied twice - 0° and 180° for each orientation8. The diffusion data were acquired in 3 shells, with q values of 84.8, 71.6, and 56.6 mm-1, each with 37, 16, and 7 orientations, respectively. AMD was estimated using a three-dimensional implementation of the MCF method9,10. To validate the MRI results immunohistochemistry for axonal neurofilament protein was performed in slices from each brain containing the optic tract11.Results and Discussion
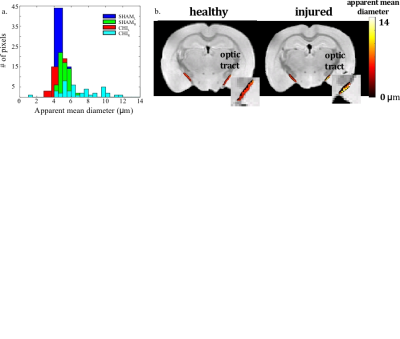
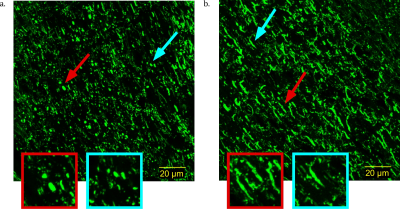
Figure 1a shows the optic tract AMD histograms from the sham and injured brains; 1b shows the optic tract AMD maps of the Sham and CHIMERA injured mice, respectively. ROI analysis of the optic tract shows the ROI average AMD of 4.8 and 5.2 μm for the right and left tracts of the Sham brain, respectively, and 8.6 and 5.4 μm for right and left tracts of the CHIMERA brain, respectively. The estimated AMD is noticeably larger in the right optic tract of CHIMERA injured mouse compared with the Sham mouse indicating an alteration in the tissue microstructure between the healthy and TBI brains. Figure 2 presents confocal images of the neurofilament protein in the optic tract. The presence of axonal varicosities, which is a characteristic pathology of mild TBI in the right tract of the injured mice verify the MRI findings11.Conclusion
DDE MRI shows promise in detecting cellular and microstructural alterations between DAI injured and healthy brains. Efforts are underway to develop it into a new quantitative imaging method with a long-term goal to detect TBI.Acknowledgements
This work was supported by the Intramural Research Program of
the Eunice Kennedy Shriver National Institute of Child Health and
Human Development [grant numbers HD000266] and The Henry
M. Jackson Foundation for the Advancement of Military Medicine,
Inc, [HJF Award Numbers: 308049-8.01-60855 and 307513-3.01-
60855].
References
1. J.M. Meythaler, et al, Current Concepts: Diffuse Axonal Injury–Associated Traumatic Brain Injury, Arch Phys Med Rehabil 82 (2001) 1461-1471.
2. M. Gaetz, The neurophysiology of brain injury, Clin Neuropathol, 115 (2004) 4–18.
3. D.R. Namjoshi, et al, Wellington, Merging pathology with biomechanics using CHIMERA (Closed-Head Impact Model of Engineered Rotational Acceleration): a novel, surgery-free model of traumatic brain injury, Mol Neurodegener, 9 (2014) 55.
4. E.B. Hutchinson, at al, Quantitative MRI and DTI abnormalities during the acute period following CCI in the ferret, SHOCK, 46 (2016) 167–176.
5. M.C. Ricciardi, et al, Trauma-Specific Brain Abnormalities in Suspected Mild Traumatic Brain Injury Patients Identifed in the First 48 Hours after Injury: A Blinded Magnetic Resonance Imaging Comparative Study Including Suspected Acute Minor Stroke Patients, J Neurotrauma, (2016) 1-8.
6.P.P. Mitra, B.I. Halperin, Effects of finite gradient-pulse widths in pulsed-field-gradient diffusion measurements, J Magn Reson, 113 A (1995 ) 94-101.
7. M.E. Komlosh, at al, Pore diameter mapping using double pulsed-field gradient MRI and its validation using a novel glass capillary array phantom, J Magn Reson, 208 (2011) 128–135.
8. M.E. Komlosh, at al, Mapping average axon diameters in porcine spinal cord white matter and rat corpus callosum using d-PFG MRI, NeuroImage, 78 (2013) 210-216.
9. D.S. Grebenkov, Laplacian Eigenfunctions in NMR. I. A Numerical Tool, Concepts Magn Reson A, 32 (2008) 277–301.
10. E. Ozarslan, et al, A general framework to quantify the effect of restricted diffusion on the NMR signal with applications to double pulsed field gradient NMR experiments, J Chem Phys, 130 (2009) 104702.
11. M. E. Komlosh, et al, Using doouble pulsed-field gradient MRI to study tissue microstructure in traumatic brain injury (TBI) A general framework to quantify the effect of restricted diffusion on the NMR signal with applications to double pulsed field gradient NMR experiments, Microporous Mesoporous Mater, 130 (2017) in press.
Figures