5351
Features extracted from diffusion-driven tensor based morphometry can serve as a specific imaging marker for Moebius Syndrome1Quantitative Medical Imaging Section, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, MD, United States, 2Medical Genomics and Metabolic Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, United States, 3Department of Computer Science, University of Maryland: College Park, College Park, MD, United States, 4Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 5Departments of Neurology and Ophthalmology, Boston Children's Hospital and Harvard Medical School, Boston, MA, United States, 6National Institutes of Health, Bethesda, MD, United States
Synopsis
Quantitative diffusion derived metrics such as fractional anisotropy (FA), and Trace of diffusion tensor (TR) have been used in many studies to assess differences between a subject group and a control group. In this study, in addition to FA and TR, we also look at morphological differences measured by diffusion-driven tensor based morphometry (DTBM). We use DTBM to extract features for use in classification of Moebius syndrome subjects, a rare birth defect characterized by paralysis or weakness of facial muscles and impairment of ocular abduction.
Introduction
Quantitative diffusion derived metrics such as fractional anisotropy (FA), and Trace of diffusion tensor (TR) have been used in many studies to assess differences between a subject group and a control group. In this study, in addition to FA and TR, we also look at morphological differences. While voxel-wise analysis of FA or MD provides insights about changes in microstructure and architecture in a structure of interest, morphological analysis provides insights about changes in the size or shape of a given structure. Morphological information can be extracted from the deformation fields that map individual images from their native space to the common space. When the Jacobian of deformation fields are used, the method is referred to as Tensor Based Morphometry (TBM)1 and it is typically performed using T1 weighted images (T1WIs). DTI has a unique ability to identify white matter pathways in regions that appear homogenous in T1WIs. Recent advancement in EPI distortion correction and robust tensor-based registration algorithm make it possible to perform TBM using DTI images which we refer to as DTBM. In this study, we perform voxel-wise statistical analysis on a subset of Moebius Syndrome (MBS) subjects and controls to identify regions that can potentially server as an imaging marker. MBS is a rare birth defect characterized by paralysis or weakness of facial muscles and impairment of ocular abduction2,3,4. We use another subset of MBS subjects to examine whether the region/voxels identified from a group analysis could serve as an imaging marker at a single subject level. We also included data from subjects with isolated congenital facial weakness (CFW) who also exhibit facial weakness, but extraocular movement are normal, to test whether the imaging marker is unique to MBS subjects.Method
Fifteen healthy volunteers (mean age: 34 years,10 female, 5 male) with no history of neurological disorders and normal MRI, and eighteen subjects diagnosed with Moebius syndrome (mean age: 30 years,13 female, 5 male) were included in this study. Five subjects had isolated VI (abducens) and VII (facial) cranial nerve involvement (classic Moebius), whereas other subjects had additional abnormalities such as limb abnormalities or mirror movements. Additional subjects with isolated CFW (mean age: 37 years, 2 females, 3 males) were also included in this study.
All participants were scanned on a Philips 3T system with 8-channel head coil. DTI dataset consisted of seven low b-values (b=0 and 50 s/mm2), and 39 volumes with maximum b-value of 1100 s/mm2. Resolution was 2mm isotropic. To correct for EPI induced distortions, the sequence was repeated for AP, PA, LR, and RL phase encoding directions. Data were processed using TORTOISE5,6,7. Control subjects’ diffusion tensors (DTs) were used to create a study-specific control template using DR-TAMAS8. FA and TR were computed from each subject’s spatially normalized DT. In addition, log of determinant of the Jacobian (LogJ) of transformations that map each individual to the control template was calculated. FA, TR, and LogJ maps were subsequently used to find regions that exhibit differences between MBS subjects and controls using FSL randomise software9 corrected for multiple comparisons using a family-wise error rate of p < 0.05. Voxels identified in the training data as being significantly different between the two groups were used as input to the Linear discriminant analysis (LDA) in R software. Controls and classic MBS subjects were used as the training dataset, and the rest of MBS subjects and CFW subjects as a test dataset.
Results
There were neither significant differences in FA
nor TR at p < 0.05 between classic MBS subjects and controls. However,
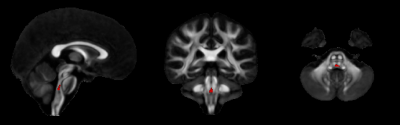
voxel-wise analysis of LogJ maps revealed volumetric reduction in the pontine
tegmentum region of the MBS subjects (Figure 1) relative to the controls.
This region is consistent with the location of the medial longitudinal
fasciculus (MLF), an important structure connecting the
nuclei of the nerves involved in ocular movements. Using DTBM and LDA algorithm
92.3% of MBS subjects were correctly classified as having MBS (12/13), and 100%
isolated CFW subjects were correctly classified as not having MBS (5/5).Discussion
Feature extraction is an important step in the application of any machine learning algorithm. Here, using DTBM, we were able to identify a potential region that can serve as an imaging biomarker for MBS subjects. It is important to note this is a region that is typically difficult to image in DTI due to distortions, however, acquisition of multiple phase encoding directions allows for correction. The LogJ maps based on DTBM method provide complementary information to the diffusion metrics that are commonly used in group analysis of DTI data. Here we show that the volume of the pontine tegmentum region can serve as an imaging marker for MBS subjects.Acknowledgements
Support for this work included funding from U01 HD079068-03 grantReferences
1. Ashburner, J., Friston, K., 2003. Morphometry. In: Frackowiak, R., Friston, K., Frith, C., Dolan, R., Friston, K., Price, C., Zeki, S., Ashburner, J., Penny, W. (Eds.), Human Brain Function, 2nd Edition. Academic Press.
2. Verzijl, H.T., van der Zwaag, B., Cruysberg, J.R. and Padberg, G.W., 2003. Möbius syndrome redefined: A syndrome of rhombencephalic maldevelopment. Neurology, 61(3), pp.327-333.
3. Webb, B.D., Shaaban, S., Gaspar, H., Cunha, L.F., Schubert, C.R., Hao, K., Robson, C.D., Chan, W.M., Andrews, C., MacKinnon, S. and Oystreck, D.T., 2012. HOXB1 founder mutation in humans recapitulates the phenotype of Hoxb1−/− mice. American Journal of Human Genetics, 91(1), pp.171-179.
4. MacKinnon, S., Oystreck, D.T., Andrews, C., Chan, W.M., Hunter, D.G. and Engle, E.C., 2014. Diagnostic distinctions and genetic analysis of patients diagnosed with Moebius syndrome. Ophthalmology, 121(7), pp.1461-1468.
5. Rohde, G. K., Barnett, A. S., Basser, P. J., Marenco, S. and Pierpaoli, C., 2004. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med 51(1), pp.103–114.
6. Irfanoglu, M.O., Modi, P., Nayak, A., Hutchinson, E.B., Sarlls, J. and Pierpaoli, C., 2015. DR-BUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions. NeuroImage, 106, pp.284-299.
7. Pierpaoli, C., Walker, L., Irfanoglu, M., Barnett, A., Chang, L.-C., Koay, C., Pajevic, S., Rohde, G., Sarlls, J. and Wu, M., 2010. TORTOISE: an integrated software package for processing of diffusion MRI data. ISMRM 19th annual meeting, Stockholm, Sweden.
8. Irfanoglu, M. O., Nayak, A., Jenkins, J., Hutchinson, E., Sadeghi, N., Thomas, C. and Pierpaoli, C., 2016. DR-TAMAS: Diffeomorphic registration for tensor accurate alignment of anatomical structures. NeuroImage 132, pp. 439–454.
9. Winkler, A. M., Ridgway, G. R., Webster, M. A., Smith, S. M. and Nichols, T. E., 2014. Permutation inference for the general linear model. Neuroimage 92, 381–397.