5342
A diffusion-matched principal component analysis (DM-PCA) based denoising procedure for high-resolution diffusion-weighted MRI1Biomedical Engineering, University of Arizona, Tucson, AZ, United States, 2University of Hong Kong, Hong Kong, Hong Kong, 3University of Arizona, Tucson, AZ, United States
Synopsis
A concern with high-resolution DWI and DTI is the limited SNR. Here we report a new denoising procedure, termed diffusion-matched principal component analysis (DM-PCA), which comprises 1) identifying a group of voxels with very similar signal variation patterns along the diffusion dimension, 2) performing PCA along the diffusion dimension for those voxels, and 3) suppressing noisy PCA components. The DM-PCA method performs reliably for input data with a range of SNR and different numbers of diffusion encoding scans, without compromising anatomic resolvability, and should prove highly valuable for imaging studies in research and clinical uses.
Introduction
A major concern with high-resolution diffusion-weighted imaging (DWI) and diffusion-tensor imaging (DTI) is the limited signal-to-noise ratio (SNR) [1,2]. Although the SNR of DWI and DTI data can be improved by denoising in post-processing, existing denoising procedures may potentially reduce the anatomic resolvability of high-resolution imaging data. Here we report an improved denoising procedure, termed diffusion-matched principal component analysis (DM-PCA), to effectively reduce noise in high-resolution DWI and DTI data without compromising anatomic resolvability.Theory
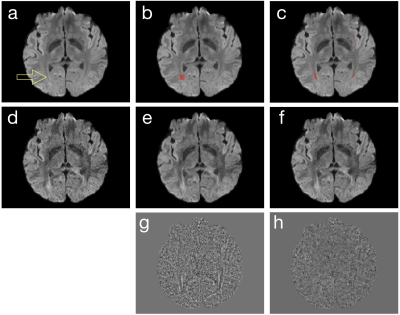
Step 1: We first identify a group of voxels, which have similar signal variation patterns along the diffusion dimension but are not necessarily nearest neighboring, from noisy input data. For example, the red dot in Figure 1a shows a target voxel, whose corresponding DWI signals are to be denoised. Instead of filtering DWI signals of the nearest neighboring voxels in a patch (Figure 1b) as implemented in the local PCA denoising method [3], we use L1 norm to identify a group of voxels (Figure 1c) that have the most similar diffusion signal variations to the target voxel.
Step 2: DWI signals of diffusion-matched voxels are processed with PCA, along the diffusion dimension using PCA, suppressing noisy PCA components. Figure 1d shows one of the 6-direction DWI input data. Using local PCA denoising (that suppresses PCA components 3 to 6 across voxels shown in Figure 1b), the produced image has a higher SNR (Figure 1e) than input data. With the DM-PCA method (that suppresses PCA components 3 to 6 across voxels shown in Figure 1c), the noise in input data can also be reduced (Figure 1f). Upon examining the residual signals of local PCA (Figure 1g) and DM-PCA (Figure 1h), it can be seen that some anatomic features are undesirably altered in local PCA produced data (likely due to the low level of redundancy in diffusion information near those voxels) but not in DM-PCA produced data.
Methods
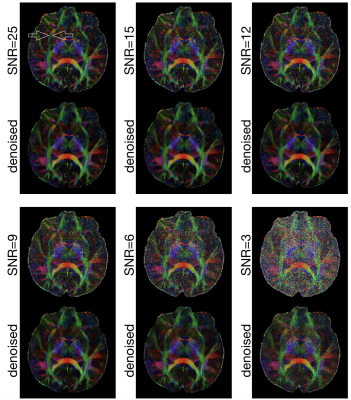
The performance of our denoising procedure was evaluated in high-resolution human brain DWI data (voxel size = 0.85 mm3, 12 diffusion-encoding directions, b = 1000 s/mm2). The SNR of this DWI dataset was ~ 25. In order to assess the performance of DM-PCA on DWI data with different SNR levels, noise was mathematically added to the data to produce five additional datasets with decreasing SNR (15, 12, 9, 6, 3).
The DM-PCA method was also applied to process 18 sets of human DTI data with conventional spatial-resolution (1.8mm3), b = 800 s/mm2 and number of diffusion-encoding directions = 25. Images were analyzed with the DTI fitting program in FSL. The sum of squared errors from DTI fitting were compared between images before and after applying the DM-PCA denoising method.
Results
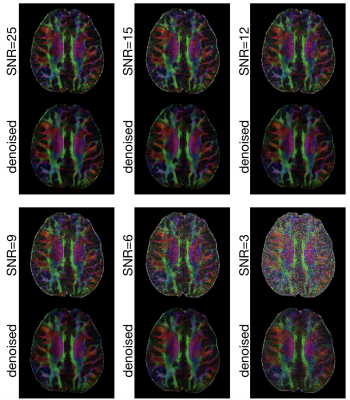
Figure 2 shows FA maps, of a chosen axial slice, calculated from high-resolution DWI data (0.85 mm3) with 6 SNR levels. The external capsule and extreme capsules are resolvable in the FA map calculated from the original DWI data (SNR ~=25) both before and after denoising. It can be seen that those two tracts remain resolvable for denoised images produced from input data with SNR = 9 or higher, demonstrating that the developed DM-PCA method does not noticeable reduce anatomic resolvability for our test datasets. Figure 3 shows the FA maps before and after applying denoising procedures to high-resolution DWI data of another axial-plane slice (arranged in the same way as that in Figure 2).
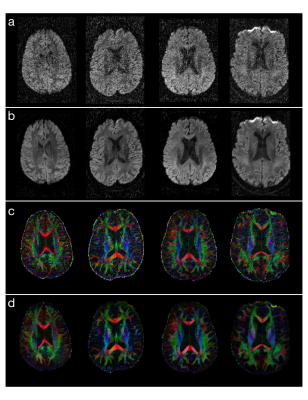
Experimental results show that the new denoising method consistently improves the SNR for all of the human DWI data (1.8 mm3 voxel size) evaluated in this study. The sum of squared errors in DTI fitting can be reduced by ~90% for all of the data obtained from 18 participants. Figures 4a and 4b compare images (corresponding to a single encoding direction) before and after applying DM-PCA, respectively, for 4 of the subjects. Figures 4c and 4d show the corresponding FA maps before and after DM-PCA processing, respectively, in which the improvement in FA quality due to denoising can be visualized.
Discussion
One of the main contributions of our study is the development of a L1-norm based procedure for identifying a group of voxels with matched diffusion properties from noisy input data. Because of the high redundancy, across the diffusion-matched voxels, on their signal variation patterns along the diffusion dimension, noise in DWI data can be reliably removed with PCA without noticeably compromising anatomic resolvability. Our experimental results suggest that the DM-PCA denoising method performs reliably on human DWI and DTI data. The DM-PCA method should prove highly valuable for high-resolution DWI and DTI studies in research and clinical uses.
Acknowledgements
This research is supported by NIH Grants R01 NS 074045 and R21 EB 018419.References
[1] Miller, K.L., Stagg, C.J., Douaud, G., Jbabdi, S., Smith, S.M., Behrens, T.E.J., Jenkinson, M., Chance, S.A., Esiri, M.M., Voets, N.L., Jenkinson, N., Aziz, T.Z., Turner, M.R., Johansen-Berg, H. and McNab, J.A. 2011. Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. Neuroimage 57(1), pp. 167–181.
[2] Chang, H.-C., Guhaniyogi, S. and Chen, N.-K. 2015. Interleaved diffusion-weighted improved by adaptive partial-Fourier and multiband multiplexed sensitivity-encoding reconstruction. Magnetic Resonance in Medicine 73(5), pp. 1872–1884.
[3] Manjón, J.V., Coupé, P., Concha, L., Buades, A., Collins, D.L. and Robles, M. 2013. Diffusion weighted image denoising using overcomplete local PCA. Plos One 8(9), p. e73021.
Figures