5255
Non-invasive assessment of glioma microstructure using VERDICT MRI with comparison to histopathology1Department of Radiology, University of Cambridge, Cambridge, United Kingdom, 2Department of Radiology, Cambridge University Hospitals NHS Foundation Trust, Cambridge, United Kingdom, 3Department of Pathology, Cambridge University Hospitals NHS Foundation Trust, Cambridge, United Kingdom, 4Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge, United Kingdom, 5Department of Clinical Neurosciences, University of Cambridge, Cambridge, United Kingdom
Synopsis
Gliomas are characterized by diffuse infiltration, high heterogeneity and poor prognosis. Imaging tumor heterogeneity may improve diagnosis and therapy planning. The Vascular, Extracellular and Restricted Diffusion for Cytometry in Tumors (VERDICT) MRI technique is a multi-compartmental model that exploits tissue microstructure. This preliminary study demonstrated the feasibility of translating VERDICT MRI in human brain imaging to investigate the microstructure of glioma with an abbreviated protocol. We demonstrated that VERDICT-derived cell size does not differ from the measured size on pathological slides and we found clear trends in LGG and HGG that may be useful to better differentiate types of glioma.
Introduction
Gliomas are characterized by diffuse infiltration and high heterogeneity, both structurally and biologically, which contributes to a very poor prognosis. The VERDICT MRI technique is a multi-compartmental model (intracellular, intravascular and extracellular–extravascular compartments) that probes tissue microstructure (1). This model has been successfully applied to xenograft models of colorectal cancer and has been translated in patients with prostate cancer (2). Here we apply VERDICT to gliomas in a clinical setting. This prospective study aimed to assess the feasibility of VERDICT in determining the microstructure of glioma in a clinical environment and explore the potential applications of VERDICT-derived parameters as imaging biomarkers to probe tumor heterogeneity, differentiate glioma subtypes and assess the peri-tumoral region.Methods
13 consecutive treatment-naive patients (5/13, 38.7% male; age 44 ± 14.7 years) with suspected gliomas underwent a DWI for VERDICT modelling, ADC was obtained for comparison. DWI for VERDICT modelling was acquired in all the patients with the following parameters: ∂ = 4.7, 12.2, 25.8, 16.5, 24.8 ms; ∆ = 23.5, 31.3, 43.4, 32.1, 43.8 ms; |G| = 49.4, 41.5, 30.1, 75.8, 43.9 mT/m – resulting in b-values of 90, 500, 1500, 2000 and 3000 s/mm2 (2). For each b-value, a separate b0, with the same number of Number of Signals Averaged (NSA) was performed. Tumor cell radii, intracellular (IC) volume and combined extracellular/intravascular volume (EC) were estimated in MATLAB 2016b (the MathWorks, Natick, MA) using a framework based on linearization and convex optimization (3,4). The maps were registered to post Gadolinium T1W and T2W sequences. A neuroradiologist outlined the regions-of-interest (ROIs) for the surrounding edema, the enhancing tumor and the necrosis on the axial T2WI and the axial 3D-T1WI post-Gd sequences. The same ROIs were applied to the corresponding registered VERDICT maps to extrapolate the microstructure parameters. A neuropathologist evaluated all the pathological slides with a semi-automated software (Image-Pro Insight, Media Cybernetics, MD) assessing cellularity and cells size. Comparisons between pathological analysis and VERDICT-derived parameters were performed using the paired sample t-test; the unpaired sample t-test was used to assess the difference in cell size and density between Low Grade Glioma (LGG) and High Grade Glioma (HGG).Results
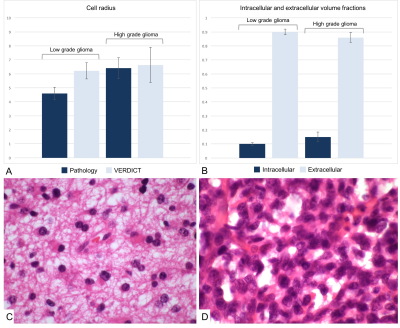
Cell radii were greater in the HGGs (6.74 ± 2.29 µm) than in the LGGs (6.22 ± 1.16; figure 1). The paired sample t-test did not show any significant difference between VERDICT-derived parameters and cell radii measured on pathological slides (p = 0.72 for HGG and p = 0.30 for LGG). The intracellular volume fraction was higher in the HGGs than in the LGGs (0.15 ± 0.07 vs 0.10 ± 0.02 respectively, p = 0.09; figure 1). The extracellular volume fraction showed an opposite trend with higher figures in LGG as compared to HGG (0.90 ± 0.04 vs 0.86 ± 0.07 respectively, p = 0.15; figure 1).Discussion
Here have applied the VERDICT MRI protocol to investigate the microstructure of gliomas in humans for the first time. There was no statistically significant difference between the average cell radii measured on pathological slides and the average VERDICT-derived parameters measured for each tumor grade (p = 0.72 for HGG and p = 0.30 for LGG). However, the cell sizes measured on pathology were slightly smaller than the ones obtained from VERDICT, which can be explained by cell shrinkage on fixation; this was confirmed by the reduction in size demonstrated for red blood cells following fixation. VERDICT-derived parameters showed a trend towards a higher intracellular volume fraction and a lower extracellular volume fraction in LGGs compared to HGGs. This can be explained pathologically as more aggressive tumors have larger cells with a higher intracellular volume fraction, whilst the extracellular volume fraction is commensurately smaller. Further work is required from larger studies to confirm this.Conclusion
This preliminary study demonstrated the feasibility of translating VERDICT MRI in brain imaging to investigate the microstructure of glioma in humans using an abbreviated protocol. We demonstrated that VERDICT-derived cell size does not differ from the measured size on pathological slides confirming that it is an accurate representation of the histopathological changes. High grade tumors showed a trend towards larger cells and a small extracellular space which is in keeping with the pathological appearances of more aggressive tumors.Acknowledgements
This study was supported by the CRUK-EPSRC Cancer Imaging Centre in Cambridge and Manchester, the NIHR Cambridge Biomedical Research Centre and the Cambridge Experimental Cancer Medicine Centre (ECMC).References
References
1. Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, et al. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res. 2014;74(7):1902–12.
2. Panagiotaki E, Chan RW, Dikaios N, Ahmed HU, O’Callaghan J, Freeman A, et al. Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Invest Radiol. 2015;50(4):218–27.
3. Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage. The Authors; 2015;105:32–44.
4. Bonet-Carne E, Daducci A, Panagiotaki E, Johnston E, Stevens N, Atkinson D, et al. Non-invasive quantification of prostate cancer using AMICO framework for VERDICT MR. Int Soc Magn Reson Med. 2016;(3):5–8.
Figures