5225
Deep Learning Captures More Accurate Diffusion Fiber Orientations Distributions than Constrained Spherical Deconvolution1Computer Science, Vanderbilt University, Nashville, TN, United States, 2Biomedical Engineering, Vanderbilt University, Nashville, TN, United States, 3Electrical Engineering, Vanderbilt University, Nashville, TN, United States, 4Biostatistics, Vanderbilt University, Nashville, TN, United States, 5Psychology, Vanderbilt University, Nashville, TN, United States
Synopsis
Confocal histology provides an opportunity to establish intra-voxel fiber orientation distributions that can be used to quantitatively assess the biological relevance of diffusion-weighted MRI models, e.g., constrained spherical deconvolution (CSD). Here, we apply deep learning to investigate the potential of single shell diffusion-weighted MRI to explain histologically observed fiber orientation distributions (FOD) and compare the derived deep learning model with a leading CSD approach. This study (1) demonstrates that there exists additional information in the diffusion signal that is not currently exploited by CSD, and (2) provides an illustrative data-driven model that makes use of this information.
Introduction
Data
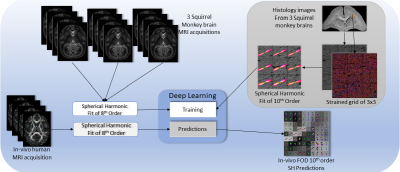
Three ex vivo squirrel monkey brains were imaged on a Varian 9.4T scanner. Briefly, data were acquired with a 3D diffusion-weighted EPI sequence (b-value=6,000 s/mm2, 100 directions) at 300um isotropic resolution. After scanning, the tissue was sectioned, stained with the fluorescent DiI, and imaged on an LSM710 Confocal microscope following the procedures outlined in [6]. The histological FOD was extracted using structure tensor analysis. Finally, a multi-step registration procedure [6] was used to determine the corresponding diffusion MRI signal. A total of 567 histological voxels were processed, and a hundred random rotations were applied to each one of them for both the MR signal and the histology FOD to augment the data bringing the total to 57267 voxels [7].
For qualitative validation, a single healthy human volunteer was scanned for a single session using a 3T (Achieva, Philips Medical Systems, Best, The Netherlands) with a 32-channel head coil. Four scans acquired were at a b-value of 2000 s/mm2 (which approximates the diffusion contrast of a fixed ex vivo scan at a b-value of 6000 s/mm2) with 96 gradient directions and an additional b0 per scan (2.5mm isotropic resolution, matrix of 96x96, 38 slices, Multi-Band=2; SENSE=2.2;TR= 2650 ms; TE=94 ms; partial Fourier=0.7). Standard pre-processing with FSL (topup, eddy correction, registration, averaging across scans) was performed before analysis.
Method
Both ex vivo and in vivo HARDI acquisitions were fit with 8th order real spherical harmonics. Outliers were manually reviewed for imaging artifacts, and 54 voxels were removed. FOD’s from the histology were fitted with a 10th order real spherical harmonics. Histology data was divided into training/validation (44,541 voxels) and testing sets (7,272 voxels) without mixing augmented data (rotations). For training/validation, a 20% percent split was used with 5 folds. Mean squared error was used to assess model accuracy [8].Results
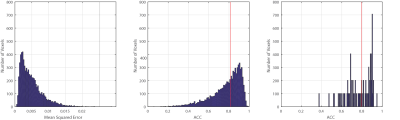
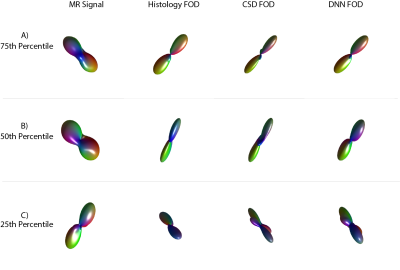
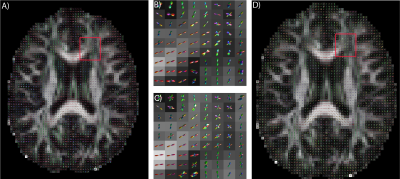
The median angular correlation coefficient (ACC) for CSD (0.7965) was significantly (p<0.05, non-parametric signed rank test) lower than for the deep approach (0.8165) (Fig 2), which corresponded to a lower root mean squared error for the deep approach (0.539 versus 0.561). Qualitatively, the predicted FOD’s on the human in vivo data demonstrate anatomical consistency (Fig 3), indicating that the deep learning approach is predicting structure in line with prior observations.Discussion
By demonstrating superiority of a deep learning approach over a leading model-based approach, CSD, we show that (1) there exists additional information in the diffusion signal that is not currently exploited by CSD, and (2) provide an illustrative data-driven model that makes use of this information. In a preliminary analysis, we applied the same network to ex vivo imaging at a b-value of 9000 s/mm2 and found a significantly higher ACC (0.850, p<0.05, non-parametric signed rank test) for deep learning which is 6.7% higher than CSD. Hence, generalizing the deep learning to use multiple shells and adapt to high b-values is a promising area of exploration. To enable others to investigate our results, the derived TensorFlow models that describe the identified MRI:histology relationships are available on the NITRC project “masimatlab” . Perhaps most importantly, this deep learning analysis demonstrates that current models for identifying fiber orientation distributions do not make all possible use of existing information, and additional innovation is possible. The deep learning models presented herein are preliminary and have not guaranteed optimality properties, and further exploration of the space of multi-layer neural networks is warranted. Additionally, continued refinement of deep learning approaches could make use of both traditional data augmentation of ground truth (e.g., rotations as used herein), but also physics/diffusion simulations of modeled geometry along with image acquisition models.Acknowledgements
Research reported in this publication was supported in part by the National Institutes of Health R01EB017230 (Landman) and R01NS058639 (Anderson). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Confocal imaging was performed in the Digital Histology Shared Resource at Vanderbilt University Medical Center (www.mc.vanderbilt.edu/dhsr)References
1.) Tournier, J-Donald, et al. "Resolving crossing fibres using constrained spherical deconvolution: validation using diffusion-weighted imaging phantom data." Neuroimage 42.2 (2008): 617-625.
2.) Anderson, Adam W. "Measurement of fiber orientation distributions using high angular resolution diffusion imaging." Magnetic Resonance in Medicine 54.5 (2005): 1194-1206.
3.)Descoteaux, Maxime, et al. "Regularized, fast, and robust analytical Q‐ball imaging." Magnetic resonance in medicine 58.3 (2007): 497-510.
4.) Jansons, Kalvis M., and Daniel C. Alexander. "Persistent angular structure: new insights from diffusion magnetic resonance imaging data." Inverse problems 19.5 (2003): 1031.
5.) Koppers, Simon, Christoph Haarburger, and Dorit Merhof. "Diffusion MRI Signal Augmentation: From Single Shell to Multi Shell with Deep Learning." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2016.
6.) Schilling, Kurt, et al. "Comparison of 3D orientation distribution functions measured with confocal microscopy and diffusion MRI." Neuroimage 129 (2016): 185-197.
7.) Krizhevsky, Alex, Ilya Sutskever, and Geoffrey E. Hinton. "Imagenet classification with deep convolutional neural networks." Advances in neural information processing systems. 2012.
8.) Tieleman, Tijmen, and Geoffrey Hinton. "Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude." COURSERA: Neural networks for machine learning 4.2 (2012): 26-31.
Figures