5217
Generation of a muscle fibre orientation atlas of the in vivo tongue1Department of Head and Neck Oncology and Surgery, Netherlands Cancer Institute/Antoni van Leeuwenhoek Hospital, Amsterdam, Netherlands, 2Department of Radiology, Academic Medical Center, Amsterdam, Netherlands, 3Department of Oral and Maxillofacial Surgery, Academic Centre for Dentistry Amsterdam and Academic Medical Center, University of Amsterdam and VU University, Amsterdam, Netherlands, 4Department of Oral and Maxillofacial Surgery, Academic Medical Center, Amsterdam, Netherlands, 5Department of Robotics and Mechatronics, MIRA Institute, University of Twente, Enschede, Netherlands, 6Department of Biomedical Engineering and Physics, Academic Medical Center, Amsterdam, Netherlands
Synopsis
While diffusion tensor imaging based atlases in the brain are common, no muscle fibre atlases for the tongue have been created. An unbiased atlas was generated from ten healthy volunteers, by iteratively registering fibre orientation distributions calculated via constrained spherical deconvolution to a template. The atlas shows good reproducibility and better performance than an image intensity based atlas. Various tongue muscles can be detected in tractography of the atlas. This atlas may facilitate automatic segmentation of tongue muscles after tumour resection to evaluate impaired tongue function.
Purpose
While diffusion tensor imaging based atlases in the brain are common1, no attempts have been performed to generate atlases for the tongue. An atlas based on muscle fibre orientation in the tongue may automate the segmentation of tongue muscles. In addition, such an atlas can be instrumental in assessing muscle function before and after surgery, which is the primary treatment option for cancer involving the oral tongue. The complex muscular architecture can be reconstructed in vivo using constrained spherical deconvolution (CSD) tractography2 of diffusion-weighted imaging (DWI). In this work, we aim to generate a tongue atlas, based on muscle fibre architecture acquired in vivo.Methods
Ten healthy volunteers were scanned in a 3T Philips Ingenia scanner (Philips Healthcare, Best, The Netherlands) using two flexible surface coils (15 cm in diameter) gently strapped to the cheeks of the volunteers. Scan parameters were: SE single-shot EPI; ETL 25; TE/TR 60ms/3.4s; two repetitions with opposing phase-encoding directions (RL&LR); NSA=1; SPIR and SSGR fat suppression; FOV 192x156x84mm; voxel size 3mm isotropic; b-value 700s/mm2 in 64 directions evenly spaced over a hemisphere and optimised for gradient load; total scan time of 10 minutes. The acquisition was repeated after repositioning the subject within an hour.
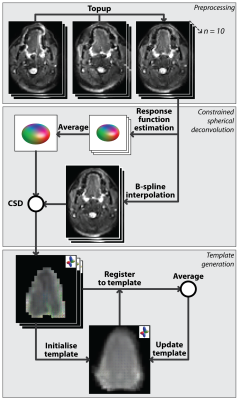
Image processing consisted of preprocessing, CSD calculation, and template generation as visualised in figure 1. Preprocessing of DWI scans consisted of denoising3 in MRtrix4, and B0-inhomogeneity, eddy current distortion, and rigid motion correction5 in FSL6. Masks of the tongues of all subjects were created by manual delineation in ITK-Snap7. All subsequent steps were performed using MRtrix4. CSD8 was performed by first estimating and single-fibre response functions9 then averaging these functions. Secondly, DWI scans were interpolated (b-spline) to a resolution of 1.5mm isotropic10, and finally fibre orientation distributions (FODs) were calculated using a maximum spherical harmonic degree ($$$l_{max}$$$) of 8.
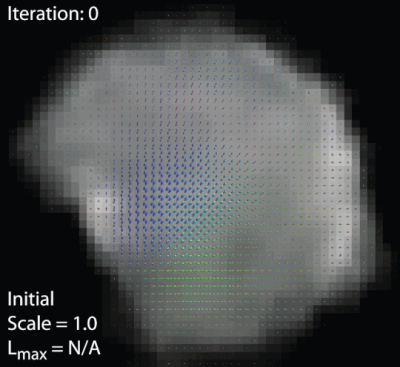
The unbiased FOD-based template of the tongue musculature was generated iteratively. An initial template was generated by averaging all 10 FOD volumes, which were aligned based on their centre of gravity. Subsequently, all FOD volumes were registered to the current template using an FOD-based registration11 with appropriate FOD reorientation12. The current template was replaced by the average of registered FOD volumes. These two steps were repeated 28 times, increasing the leniency of the registration with each step (figure 2). To assess reproducibility, the complete processing pipeline was repeated using the second acquisitions. Finally, to determine the improvement of FOD-based registration compared to image intensity based registration, a DWI-based template was generated similarly by iteratively registering averaged b0-images instead of FODs. Finally, FODs were calculated from the DWI-based template using the averaged response function.
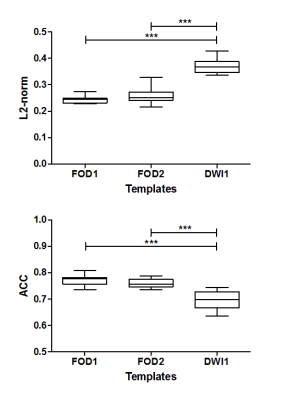
Template quality was assessed by calculating the similarity on a voxel-level of FODs between the ten subjects transformed to the template and the template. Two metrics, namely the spherical harmonic L2-norm (L2) and angular correlation coefficient (ACC)12 were calculated and averaged over the whole tongue. For the DWI-based template, DWI scans were registered to the DWI-based template and deconvolved into FOD volumes, and mean L2 and ACC were calculated. Two-tailed paired t-tests were performed on both mean L2 and ACC (5% significance level).
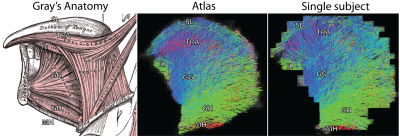
CSD-based fibre tractography13 was performed on subject-specific FOD volumes and the final template in order to create a fibre atlas, with the following parameters: step size 1.5mm; angular threshold 15°; maximal length 100mm; minimal length 10mm; 10000 seed point randomly placed with the mask.
Results
Figure 3 shows the similarity between all subject-specific FOD volumes and the corresponding template. FOD-based templates display significantly better similarity in both repetitions, compared to the DWI-based template. The templates from both repetitions were not significantly different. Tractography of the atlas (figure 4) displays several tongue muscles in agreement with the anatomical atlas, and performs better than subject specific tractography.Discussion
For the first time, an atlas based on muscle fibre directionality measured by DWI has been created of the in vivo human tongue. Tongue atlases have been reported using serial sectioning14 or T2-weighted MRI15. However, these methods are either based on ex vivo data or do not explicitly take into account fibre directionality, which is essential for the complex muscular architecture of the tongue. This tongue atlas may facilitate automated segmentation of tongue muscles in single subjects to evaluate impaired tongue function after tumour resection.Acknowledgements
We are grateful to the developers of the open-source software toolbox MRtrix.References
1. Mori S, Oishi K, Faria A. White matter atlases based on diffusion tensor imaging. Curr Opin Neurol. 2009;22(4):362-369.
2. Voskuilen L, Mazzoli V, Oudeman J, et al. Crossing muscle fibres in the tongue resolved using constrained spherical deconvolution. In: Proc of ISMRM, Honolulu, Hawaii. 2017.
3. Veraart J, Novikov D, Christiaens D, Ades-aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. Neuroimage. 2016;142:1-28.
4. Tournier J, Calamante F, Connelly A. MRtrix: Diffusion tractography in crossing fiber regions. Int J Imaging Syst Technol. 2012;22(1):53-66.
5. Andersson J, Sotiropoulos S. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016;125:1063-1078.
6. Smith S, Jenkinson M, Woolrich M, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(SUPPL. 1):208-219.
7. Yushkevich P, Piven J, Hazlett H, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116-1128.
8. Tournier J, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage. 2007;35:1459-1472.
9. Tournier J, Calamante F, Connelly A. Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed. 2013;26(12):1775-1786.
10. Dyrby T, Lundell H, Burke M, et al. Interpolation of diffusion weighted imaging datasets. Neuroimage. 2014;103:202-213.
11. Raffelt D, Tournier J, Fripp J, Crozier S, Connelly A, Salvado O. Symmetric diffeomorphic registration of fibre orientation distributions. Neuroimage. 2011;56(3):1171-1180.
12. Raffelt D, Tournier J, Crozier S, Connelly A, Salvado O. Reorientation of fiber orientation distributions using apodized point spread functions. Magn Reson Med. 2012;67(3):844-855.
13. Tournier J, Calamante F, Connelly A. Improved probabilistic streamlines tractography by 2nd order integration over fibre orientation distributions. ISMRM. 2010;88(2003):2010.
14. Sanders I, Mu L. A three-dimensional atlas of human tongue muscles. Anat Rec. 2013;296(7):1102-1114.
15. Woo J, Lee J, Murano E, et al. A High-resolution Atlas and Statistical Model of the Vocal Tract from Structural MRI. Comput Methods Biomech Biomed Eng Imaging Vis. 2015;3(1):47-60.
Figures