5211
Personalized Lesion Profiling in Multiple Sclerosis1Brain and Mind Centre, University of Sydney, Sydney, Australia, 2Save Sight Institute, University of Sydney, Sydney, Australia, 3Faculty of Medicine and Health Sciences, Macquarie University, Sydney, Australia, 4Sydney Neuroimaging Analysis Centre, Sydney, Australia, 5School of Instrumentation Science and Opto-Electronics Engineering, Beihang University, Beijing, China, 6School of Information Technologies, University of Sydney, Sydney, Australia
Synopsis
The pathology of multiple sclerosis (MS) lesions is highly subject-specific, therefore personalized lesion analysis is important in assessing the degree of disease-related brain damage and the patient’s response to therapy. Separating and comparing lesional and non-lesional tissue provides an “internal control” and, therefore, increased precision of measurement of the degree of lesional damage in the brain. Here we present a novel and fully automatic method to create a personalized lesion profile of the subject based on DTI tractography, which can be used to assess whole brain lesional damage. This technique can be applied to measure microstructural change using any MRI modality, and may have high translational impact in clinical trials and neurological research.
Introduction
The pathology of Multiple Sclerosis (MS) lesions is highly subject-specific, therefore personalized analysis is important in early diagnosis and treatment planning for MS patients [1]. Conventional MRI techniques provide no or little pathology-specific MS lesion information.
Studies on the asymmetry between lesional and non-lesional tissues in the brain may provide a precise measure to quantify the degree of lesional damage by using non-lesional tissue as an internal control [2]. We previously proposed a method to measure the microstructural changes in lesion and non-lesional fibres along the same white matter pathway, using optic radiation (OR) as an example [3]. This analysis, however, was limited to highly coherent pathways, required manual selection of the fibre bundles to exclude crossing fibres, and further pruning of false fibre branches.
Here, we present a novel and fully automatic method to identify the lesional and non-lesional fibre tracts in whole brain, and create a personalized lesion profile. This technique can be used to measure the microstructural changes using any MRI sequence, such as T1, FLAIR, DTI and MTR, and may have potential applications in clinical trials and the study of MS lesion dynamics.
Methods
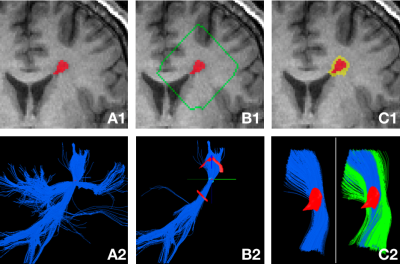
The proposed Personalized Lesion Profiling (PLP) technique is based on DTI tractography. Step 1: Deterministic tractography was performed to reconstruct the white matter tracts using individual lesions as seeding points, as shown in Figure 1 (A1 – A2). Step 2: A peri-lesional shell was created by expanding the lesion boundary by 15 voxels (which can be defined by the user), as shown in Figure 2 (B1). The shell was then intersected with the fibre tracts to identify the traversing voxels of the fibres on the shell, which can be clustered into different patches, as shown in Figure 2 (B2). Fibres were grouped into the same fibre bundle if they traversed the same patches, otherwise into separate fibre bundles. Figure 1 (B2) shows that a single coherent fibre bundle was identified, whose fibres all passed through the lesion and the same inbound and outbound traversing patches. Step 3: The lesion mask was dilated by 2 voxels (which can be defined by the user) to create the peri-lesional mask, which was then used to derive the non-lesional fibres. Figure 1 (C2) shows the lesional (blue) and non-lesional fibres (green), which were further clipped using the shell.
The previous steps were repeated for individual
lesions in the brain, resulting in the same number of pairs of lesional and
non-lesional fibres as the lesions. Figure 2 shows an example of the whole
brain lesional and non-lesional fibre tracts grouped in pairs for individual
lesions. The tracts were then mapped to a parametric
image, such as the functional parametric images derived from DTI and MTR, to
measure the microstructural changes along the tracts.
Results
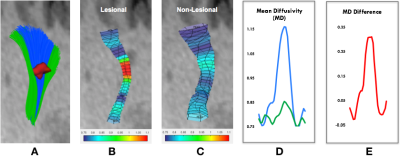
We used the mean diffusivity (MD) map derived from DTI as an example to demonstrate the use of PLP technique. For each pair of lesional and non-lesional fibres, the average weighted MD values were calculated at every cutting point with 1 mm intervals (which can be defined by the user) along the tracts. The non-lesional MD values were then subtracted from the lesional MD values at corresponding positions to normalize the lesional profile. Figure 3 (A-C) shows an example of the reconstructed lesional (blue) and non-lesional fibres (green), and the MD values along them, respectively. Figure 3 (D) shows plots of the lesional and non-lesional MD values on the same scale. Figure 3 (E) shows the normalized lesional profile.
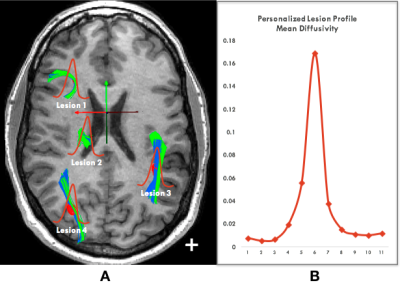
The normalized profiles of all lesions in
the brain were further averaged into a single patient-based profile
(which we called personalized lesion
profile), as shown in Figure 4.
Conclusion
This study represents the first attempt to derive a whole brain based personalized lesion diffusivity profile. Similar profiles can be constructed for other modalities (e.g., MTR). Such personalized profiles will enable the comparison between different subjects and populations, and may have high translational impact in clinical trials and neurological research.Acknowledgements
No acknowledgement found.References
[1] A. Klistorner, C. Wang, C. Yiannikas, S.L. Graham, J. Parratt, M.H. Barnett, "Progressive Injury in Chronic Multiple Sclerosis Lesions is Gender Specific: A DTI Study", PLOS ONE 11(2): e0149245, 2016.
[2] A. Klistorner, C. Wang, C. Yiannikas, M. Barnett, J. Parratt, "Accelerated Loss of Chronically Demyelinated Axons in Relapsing-Remitting Multiple Sclerosis", ECTRIMS: 675, 2017.
[3] A. Klistorner, N. Vootakuru, C. Wang, C. Yiannikas, S.L. Graham, J. Parratt, R. Garrick, N. Levin, L. Masters, J. Lagopoulos, M.H. Barnett, "Decoding Diffusivity in Multiple Sclerosis: Analysis of Optic Radiation Lesional and Non-Lesional White Matter", PLOS ONe 10(3): e0122114, 2015.
Figures