4897
Self-gated respiratory-resolved 5D Flow MRI using the 3D spiral phyllotaxis trajectory1CNRS, University of Lyon, CREATIS Laboratory, Lyon, France, 2School of Biomedical Engineering and Imaging Sciences, King’s College London, London, United Kingdom, 3Philips Healthcare, Guildford, United Kingdom, 4MR Clinical Science, Philips Healthcare, Best, Netherlands, 5Department of Interventional Radiology and Cardio-vascular and Thoracic Diagnostic Imaging, Hospices Civils de Lyon, Lyon, France
Synopsis
Clinical diagnosis based on thoracic blood flow using three-dimensional cardiac-resolved (4D) phase contrast PC-MRI is limited by long acquisition times, due to the need of respiratory gating. In addition, gating does not provide potentially important physiological information, the variation of flow within the respiratory cycle. To address these challenges we developed a 5D (4D respiratory-resolved) PC-MRI sequence combining 3D radial k-space sampling, respiratory self-gating, and compressed sensing reconstruction. Our preliminary results demonstrate respiratory related changes in blood flow in healthy subjects. The proposed method may potentially refine diagnosis in congenital heart disease by assessing the respiratory related blood flow variations.
INTRODUCTION
Clinical assessment of thoracic blood flow using three-dimensional cardiac-resolved (4D) phase contrast MRI (PC-MRI) has been limited by long acquisition times, due in part to the need of respiratory gating. Standard pencil beam respiratory gating is highly inefficient since data is only acquired during a specific phase of the respiratory cycle, leading to prolonged scan times. Moreover, gated acquisitions do not provide potentially important physiological information on the variation of flow with the respiratory cycle.
Respiratory self-gating has the potential to enable velocity measurements at different phases of the respiratory cycle without any data rejection. Our goal was to develop a respiratory self-gated 3D radial 5D (4D respiratory-resolved) flow MRI sequence and study the respiratory related variability of blood flow velocity in the thoracic aorta.
METHODS
We implemented a 3D radial trajectory based on a spiral phyllotaxis pattern for k-space sampling similar to that proposed by Piccini et al.1 on a 1.5T Philips Ingenia system (Philips, Best, The Netherlands). The trajectory has several advantages for flow imaging: minimal eddy currents compared to other 3D radial sampling patterns, intrinsic self-navigation, and isotropic resolution. We combined the trajectory with bipolar gradients on 3 fixed spatial directions for velocity encoding.
Imaging was performed on 3 healthy subjects using the implemented 3D radial sequence and a standard Cartesian 4D flow acquisition. The Cartesian acquisition was performed using pencil beam respiratory gating and prospective cardiac triggering in an oblique plane encompassing the thoracic aorta with the following parameters: TFE factor 2, turbo direction Y, k-t BLAST factor 8, TE/TR = 1.9/4.3 ms, flip angle = 6◦, VENC = 240 cm/s; FOV = 320x320x100 mm3, 2.5 mm isotropic voxel size, bandwidth = 723 Hz. The 3D radial acquisition was free-running interleaved (8 projections per interleaf) and performed with the following parameters: TE/TR 2.5/6.0 ms, flip angle = 6◦, VENC = 240 cm/s, FOV = 340x340x340 mm3, 2.5 mm isotropic voxel size, bandwidth = 723 Hz. A total of 54096 radial readouts were acquired in 6762 interleaves. Each projection was repeated 4 times for velocity measurements using Hadamard encoding.
The respiratory self-gated (SG) signal was derived independently for each velocity encoding step as the time variation of the z coordinate of the center of mass of the image2, computed from the first projection of each interleaf using a conjugate gradient method with L1 regularization. Using the respiratory SG signal, data was separated in 3 respiratory phases with equal number of projections per bin. Subsequently, each respiratory phase was binned in 8 cardiac phases using the ECG signal. Respiratory- and cardiac-resolved images were reconstructed offline from undersampled data (R ~ 9) using a compressed sensing3 algorithm implemented in Matlab (The Mathworks, Inc, Natick, MA) using first order finite differences as sparsifying transforms in space ($$$\triangledown_{s}$$$) and time ($$$\triangledown_{t}$$$), for both cardiac and respiratory dimensions:
$$\hat{I_{b}}=arg min_{I_{b}}\left\{\parallel EI_{b} - K_{b}\parallel_2^2 + \lambda_{s}\parallel \triangledown_{s}I_{b}\parallel_{1} + \lambda_{c}\parallel \triangledown_{t}I_{b}\parallel_{1}+ \lambda_{r}\parallel \triangledown_{t}I_{b}\parallel_{1}\right\}$$
where $$${I_{b}}$$$ is a temporally resolved image at a given cardiac and respiratory phase, $$$E$$$ is the encoding operator, $$$K_{b}$$$ is the undersampled k-space data and $$$ \lambda_{s}, \lambda_{c}, \lambda_{r}$$$ are regularization weights.
RESULTS
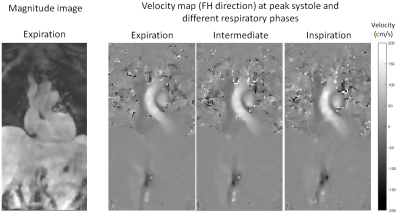
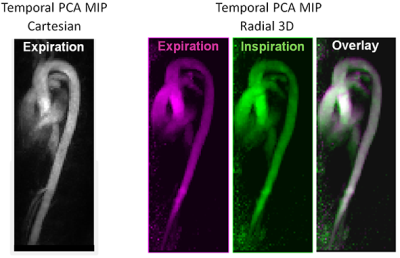
All datasets were reconstructed successfully with minimal residual breathing artefacts. An average respiratory amplitude of 9.4 ± 3.3 mm was observed. Time averaged velocity values in the thoracic aortas were 30.3 ± 3.5 cm/s for the Cartesian measurement in expiration, and varied between respiratory phases for the 3D radial measurement: from 25.0 ± 2.6 cm/s in end-expiration to 27.3 ± 6.8 cm/s in end-inspiration. Representative cardiac, respiratory and velocity resolved images acquired with the new 3D radial sequence are presented in Figure 1. Displacement of the ascending aorta due to respiratory motion can be seen in Figure 2, representing temporal PC Angiography MIPs of the end-inspiration and end-expiration phases.DISCUSSION
We present a respiratory and cardiac resolved 5D Flow MRI sequence based on a 3D radial trajectory. Our preliminary results demonstrate respiratory cycle related velocity variations. Lower overall velocity values were obtained with the 3D radial sequence compared to the Cartesian sequence, most likely as a result of temporal smoothing from the compressed sensing reconstruction. Optimization of the regularization weights controlling the tradeoff between sparsity and data consistency is ongoing to reduce this effect.CONCLUSION
Blood circulation in healthy individuals is a result of three physiological pumps: heart, peripheral muscles, and respiration. In patients suffering from congenital heart disease following corrective surgery (cavopulmonary shunt, Fontan circulation) the impact of respiration on blood flow can be significant due to strong dependence on changes in intrathoracic pressure4. Our method has the potential to refine diagnosis in congenital heart disease by assessing the respiratory related change in blood velocities.Acknowledgements
No acknowledgement found.References
[1] D. Piccini et al, “Spiral phyllotaxis: the natural way to construct a 3D radial trajectory in MRI”, Magnetic Resonance in Medicine 66:1049–1056 (2011)
[2] O. Wieben et al, “Correcting for Translational Motion in 3D Projection Reconstruction”, Proc. Intl. Soc. Mag. Reson. Med 9 (2001) #737
[3] Lustig, Michael, et al., “Sparse MRI: The application of compressed sensing for rapid MR imaging.” Magn. Reson. Med. 2007; Vol. 58:1182-1195
[4] H. Korperich et al, “Differentiation of impaired from preserved hemodynamics in patients with Fontan circulation using real-time phase-velocity cardiovascular Magnetic Resonance”, J Thorac Imaging, 32 (2017) 159-168
Figures