4881
Spin Echo Based Cardiac Diffusion Tensor Imaging of the Unfixed, Ex-vivo Porcine Heart at 7T and 3T1Chair of Cellular and Molecular Imaging, Comprehensive Heart Failure Center (CHFC), University Hospital, Wuerzburg, Germany, 2Institute of Diagnostic and Interventional Radiology, University Hospital, Wuerzburg, Germany, 3Translational Center Regenerative Therapies (TLC-RT), Fraunhofer Institute for Silicate Research (ISC), Wuerzburg, Germany
Synopsis
7T, 3D, whole heart, high resolution, diffusion tensor data sets with 1.3mm isotropic voxels, unachievable in human in-vivo scans, were acquired for 8 ex-vivo pig hearts using a Stejskal-Tanner sequence with varying parallel imaging factors. ADC, FA, helix angle and secondary eigenvector angle values were analyzed and compared to a reference set of 26 ex-vivo hearts measured with the same protocol at 3T. Purpose of this study was quality assessment in DTI of the ex-vivo porcine heart at 7T. The proof of principle data acquired allows future optimization of acquisition approaches and helps translating the method to an in-vivo application.
Introduction
Myocardial microarchitecture is remodeled in a broad range of cardiovascular pathologies. While cardiac diffusion tensor imaging (cDTI) aims to provide information on these structural changes, the technique is limited by low signal-to-noise-ratio (SNR). MRI at higher field strength such as 7T may provide higher SNR, allowing for improvement of spatial and angular resolution, b-values, image quality as well as consistency of diffusion metrics. It remains unclear, if the field-strength related gain in SNR outweighs the shorter T2, T2* and increased B0 and B1 inhomogeneity. Purpose of this study was the assessment of quality in DTI of the unfixed, ex vivo porcine heart at 7T compared to a 3T reference1 regarding susceptibility induced distortions, SNR and derived diffusion parameters for varying parallel imaging factors.Methods
Measurements were performed on 7T and 3T whole-body MRI systems (Siemens MAGNETOM™ Terra and Prisma, respectively) using a 1Tx/32Rx head coil. Admission by the institutional animal care committee was obtained before the study. Porcine hearts were stored and imaged in physiological saline solution. MRI with spatial resolution of 1.3x1.3x1.3mm3 was performed within 10 hours after cardiac arrest. 26 and 8 cDTI datasets were acquired using a Stejskal-Tanner preparation at 3T and 7T, respectively. Further measurement parameters were TE: 55ms, echo train length: 55, FOV:111x170mm2 and 5/8 partial-Fourier. 30 diffusion directions (b=700 s/mm2) and 5 b =0 s/mm2 images after Skare2 were gathered in nine minutes. The scan was repeated with GRAPPA acceleration factors R=2 (GRAPPA2) (n=4 hearts), GRAPPA3 and GRAPPA4 (n=8), and 6/8 partial-Fourier resulting in reduced echo train lengths of 33, 21 and 16. Post processing was done using DSI studio3 and MATLAB (MathWorks, Natick, USA). The myocardium was manually segmented according to the American Heart Association (AHA) model4. ADC, FA, helix angle5,6 (HA) and secondary eigenvector angle6 (E2A) were calculated and compared to the reference scan at 3T using a Mann-Whitney U test (p=0.05).Results
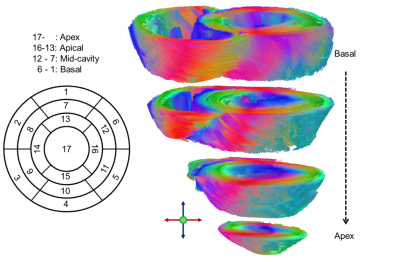
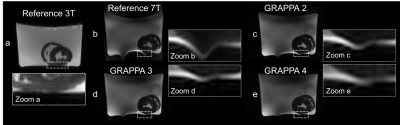
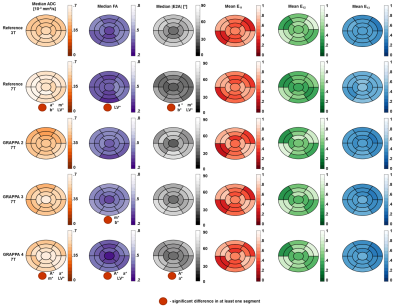
Figure 1 shows an example of the segment distribution and the main eigenvector orientation for reconstructed fibers of the left ventricle. The tradeoff between reduction of susceptibility induced distortions and the loss in SNR for increasing GRAPPA factors is displayed in Figure 2. Distortions at the container boundaries are decreasing with higher acceleration. SNR in b0 images of reference scans at 3T and 7T was ~32 and ~42. B1 variation is apparent in 7T acquisitions, but negligible for DTI of myocardial tissue. Figure 3 shows the AHA segment distribution of median ADC [10-3 mm2/s], FA, |E2A| [°], and the mean of main eigenvector (E1) components. With relative errors ranging from 17-48% for ADC and 41-55% for |E2A|, most segments significantly deviate from the reference for GRAPPA4 scans at 7T. Significant deviations (6-24%) regarding FA exist for the reference, GRAPPA3, and GRAPPA4 acquisitions at 7T. There were no significant differences in median values of ADC, FA, HA and |E2A| for measurements using GRAPPA2. Variations of up to 90% were found for mean HA values in in the apex and apical segment in the 7T reference scan. Main eigenvector components based on this scan strongly deviate from the 3T reference, while E1 distributions for all other scans at 7T were similar.Discussion
Without taking into account physiological tissue parameters the SNR gain at 7T was lower than theory would suggest7. This is most likely explained by shortened T2 and T2* relaxation times at 7T. Significant differences in ADC and |E2A| in the reference scan at 7T confirm a need for parallel imaging in order to minimize effects of geometrical distortions as shown in preliminary studies8,9. Higher acceleration, such as GRAPPA4, results in SNR penalty, which leads to significant bias in ADC, FA, and |E2A|. However the penalty is less severe for main eigenvector components. This is probably caused by the high number of diffusion directions10 (30), which balances the loss of SNR in accelerated scans and reduces hardware induced errors.Conclusion
We acquired cDTI data of ex vivo porcine hearts with a spatial resolution of 1.3x1.3x1.3mm3 and varying GRAPPA factors at 7T and compared derived diffusion metrics to a 3T reference set consisting of 26 such hearts1. ADC, FA, HA, |E2A| and other parameters were consistent with the 3T reference using GRAPPA2 acceleration. B1 inhomogeneity and the resulting areas of minimized SNR gain in 7T acquisitions are difficult to quantify and may be overcome using multichannel transmit approaches in the future. The acquired data gives information for the optimization of future ex vivo and in vivo cDTI at 7T acquisitions.Acknowledgements
Financial support: German Ministry of Education and Research (BMBF, grants: 01EO1004, 01E1O1504).References
1. Lohr D, Terekhov M, Weng A M, et al. High Resolution Cardiac DTI - Fiber Tractography Statistics of the ex Vivo pig Heart. Proceedings of the Twentyfift Annual Meeting of the International Society of Magnetic Resonance in Medicine. Honolulu; 2017. P. 1841.
2. Skare S, Hedehus M, Moseley M E, et al. Condition Number as a Measure of Noise Performance of Diffusion Tensor Data Acquisition Schemes with MRI. Journal of Magnetic Resonance. 2000; 147(2): p. 340-352.
3. Yeh F-C. DSI-Studio. http://dsi-studio.labsolver.org. Accessed June 22, 2016.
4. Cerqueira M D , Weissman N J, Dilsizian V, et al. Standardized Myocardial Segmentation and Nomenclature for Tomographic Imaging of the Heart. A Statement for Healthcare Professionals From the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. 2002; 105(4): p. 539-542.
5. Ferreira P F, Kilner P J, McGill L A, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson, 2014. 16: p. 87.
6. Mekkaoui C, Huang S, Chen H H, et al. Fiber architecture in remodeled myocardium revealed with a quantitative diffusion CMR tractography framework and histological validation. J Cardiovasc Magn Reson. 2012; 14(1).
7. Vlaardingerbroek M T, den Boer J A. Magnetic Resonance Imaging: Theory and Practice Berlin; Heidelberg: Springer; 1996. p.256.
8. Sammet S, Koch R, Irfanoglu OM, et al. SENSE factor optimization for diffusion tensor imaging of the brain at 7T. Proceedings of the Fifteenth Annual Meeting of the International Society of Magnetic Resonance in Medicine. Berlin; 2007.
9. Morgan PS, Coxon RJ, Habib J, et al. Comparison of sequences for improved diffusion weighted imaging at 7T. Proceedings of the Seventeenth Annual Meeting of the International Society of Magnetic Resonance in Medicine. Honolulu; 2009. P. 1807.
10. McClymont D, Teh I, Whittington H, et al. Evaluation of diffusion directions vs averages for accurate and reproducible cardiac DTI. Proceedings of the Twentyfift Annual Meeting of the International Society of Magnetic Resonance in Medicine. Honolulu; 2017. eP. 3352.
Figures