4879
Validation of Diffusion Tensor Imaging in Diseased Myocardium1Leeds Institute of Cardiovascular and Metabolic Medicine, University of Leeds, Leeds, United Kingdom, 2Division of Cardiovascular Medicine, Radcliffe Department of Medicine, University of Oxford, Oxford, United Kingdom, 3Diamond Light Source, Didcot, United Kingdom, 4Department of Physics and Astronomy, University College London, London, United Kingdom, 5University of Manchester, Manchester, United Kingdom, 6Feinberg School of Medicine, Northwestern University, Chicago, IL, United States
Synopsis
Diffusion tensor imaging (DTI) has been used in clinical research to identify microstructural changes in the heart following disease. However, the evidence supporting its use in the presence of abnormalities such as scar, hypertrophy and collagen infiltration is limited. Here, we investigate the application of DTI in fixed healthy, infarcted, hypertrophic and fibrotic mouse hearts, and validate these on a voxel-wise basis with high-resolution structure tensor synchrotron radiation imaging. Our findings show good agreement in helix angle estimation between the two imaging modalities across all hearts, and supports the clinical role of DTI in the presence of cardiac pathologies.
Introduction
Diffusion tensor imaging (DTI) is increasingly used to assess the 3D tissue microstructure in the heart. It is understood from prior validation studies using histology,1,2 anatomical MRI,3 CT4 and synchrotron radiation imaging (SRI),5 that the principal orientations of cardiomyocytes, sheetlets and sheetlet-normals generally correspond to the principal eigenvectors v1, ν2 and ν3 of the diffusion tensor (DT). These studies were however based on healthy fixed hearts, and did not account for potentially important cellular perturbations in diseased hearts, such as those arising from scar formation, collagen infiltration and changes in cell size and distribution. In this study, we investigate the microstructure of mouse hearts in four different states of health with DTI, including control, infarcted, hypertrophic and fibrotic hearts, and validate the results against structure tensor synchrotron radiation imaging (STSRI).Methods
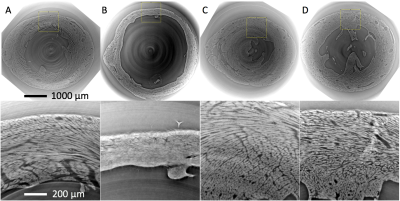
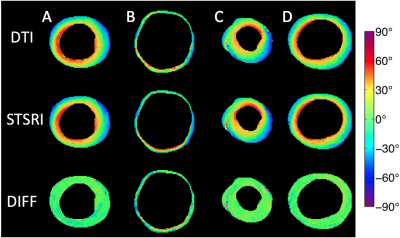
Hearts were excised from four groups of mice (n=1): healthy wild-type, two weeks after myocardial infarction (induced by transient ligation of the left anterior descending coronary artery), hypertrophic following transverse aortic constriction for 3 weeks (all C57BL/6J), and fibrotic (MLP-KO). Experimental investigations conformed to the UK Home Office guidance on the Operations of Animals (Scientific Procedures) Act 1986 and were approved by the University of Oxford ethical review board. The hearts were fixed in 4% then 1% paraformaldehyde and embedded in agarose gel for DTI and SRI. Non-selective 3D fast spin echo DTI data were acquired on a 9.4 T preclinical MRI scanner (Agilent, CA, USA) with a transmit-receive birdcage coil (Rapid Biomedical, Rimpar, Germany) of inner diameter = 20 mm. TR / TE = 1500 / 9.2 ms, echo spacing = 4.1 ms, echo train length = 8, isotropic resolution = 94 μm, number of non-DW images = 3, number of DW directions = 21, b = 1,000 s/mm2. SRI was subsequently performed at beamline I13-2 (imaging branch) of the Diamond Light Source (Didcot, UK) using an X-ray beam of 25 keV, selected by a multilayer monochromator. Data were acquired with effective pixel size = 1.1 μm, and reconstructed using a single-distance phase retrieval algorithm.6 DTs were fitted to the DTI data using non-linear least squares, while structure tensors (STs) were calculated based on the signal gradient intensity in the SRI phase contrast data.7 Data were resampled to 50 μm isotropic resolution, and manually registered. Helix angle (HA) maps were calculated from the primary eigenvector of the DT (ν1,DT) and the tertiary eigenvector of the ST (ν3,ST).5 Bland-Altman analysis of HA was performed in a single apical slice, using a mask of the left ventricular myocardium eroded by one pixel. Data were analysed in Matlab R2016A (Mathworks, Natick, USA).Results
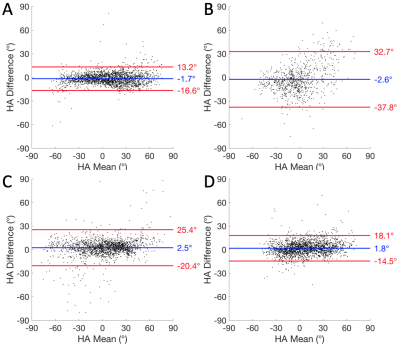
SRI phase contrast images show high fidelity of cellular structures (Figure 1). Good correspondence is observed in the HA maps calculated from the DTI and STSRI data (Figure 2). The HA differences between the two modalities in the control, infarcted, hypertrophic and fibrotic hearts were -1.6° ± 7.6°, -2.6° ± 18.0°, 2.5° ± 11.7° and 1.8° ± 8.3° respectively (mean ± SD across voxels in left ventricular myocardium; Figure 3).Discussion
DTI enables inference of the 3D orientation of tissue microstructure in the heart. With DTI finding increased application in clinical research, including those involving patients with infarct,8 hypertrophic cardiomyopathy,9 fibrosis10 and others, it is crucial that its use in the presence of such pathologies is validated. We observed that the accuracy and precision of the DTI HA estimation, with respect to the STSRI reference data, was nearly identical in the control and fibrotic hearts. There was a ~35% improvement in precision compared with prior findings in fixed healthy rat heart,5 stemming from further optimisation to the data acquisition and post-processing. The accuracy in HA estimation was good and the absolute mean difference < 3° in all cases. The precision was poorer particularly in the infarcted heart. This would have arisen from an enhancement of the effects of partial volume, cell dispersion and imperfect registration due to the thinner myocardium.Conclusion
STSRI is ideally placed for validation of DTI, as it allows for imaging of the whole rodent heart, in the same preparation as in the DTI scan. Prior evaluation of DTI in infarct11 and fibrosis12 by histology averaged over broad regions of myocardium. To our knowledge, this is the first voxel-wise validation of DTI in diseased hearts.Acknowledgements
This work was supported by the EPSRC, UK (EP/J013250/1), the BHF, UK (PG/13/33/30210; RG/13/8/30266; SI/14/1/30718), the BHF Centre for Research Excellence, UK (FS/11/50/29038; RE/13/1/30181) and the Wellcome Trust, UK (090532/Z/09/Z). We thank Dr Irene Zanette for her expert advice on SRI, and Diamond Light Source for access to beamline I13-2 (MT15287-1).References
1. Holmes AA, Scollan DF, Winslow RL. Direct histological validation of diffusion tensor MRI in formaldehyde-fixed myocardium. Magn Reson Med. 2000;44:157–61.
2. Kung GL, Nguyen TC, Itoh A, et al. The presence of two local myocardial sheet populations confirmed by diffusion tensor MRI and histological validation. J Magn Reson Imag. 2011;34(5):1080-1091.
3. Bernus O, Radjenovic A, Trew ML, et al. Comparison of diffusion tensor imaging by cardiovascular magnetic resonance and gadolinium enhanced 3D image intensity approaches to investigation of structural anisotropy in explanted rat hearts. J Cardiovasc Magn Reson. 2015;17(1):31.
4. Ni H, Castro SJ, Stephenson RS, et al. Extracting myofibre orientation from micro-CT Images: an optimisation study. In: Proceedings of the 40th Annual Conference on Computing in Cardiology, Zaragoza, Spain, 2013.
5. Teh I, McClymont D, Zdora MC, et al. Validation of diffusion tensor MRI measurements of cardiac microstructure with structure tensor synchrotron radiation imaging. J Cardiovasc Magn Reson. 2017;19:31.
6. Paganin D, Mayo SC, Gureyev TE, et al. Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. J Microsc (Oxford). 2002;206:33–40.
7. Knutsson H. Representing local structure using tensors. Linkoping: Linkoping University, Computer Vision Laboratory; 1989.
8. Wu MT, Tseng WY, Su MY, et al. Diffusion tensor magnetic resonance imaging mapping the fiber architecture remodeling in human myocardium after infarction: correlation with viability and wall motion. Circulation. 2006;114:1036–45.
9. Ferreira PF, Kilner PJ, McGill LA, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2014;16:87.
10. Nguyen C, Lu M, Fan Z, et al. Contrast-free detection of myocardial fibrosis in hypertrophic cardiomyopathy patients with diffusion-weighted cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2015;17:107.
11. Mekkaoui C, Huang S, Chen HH, et al. Fiber architecture in remodeled myocardium revealed with a quantitative diffusion CMR tractography framework and histological validation. J Cardiovasc Magn Reson. 2012;14:70.
12. Abdullah OM, Drakos SG, Diakos NA, et al. Characterization of diffuse fibrosis in the failing human heart via diffusion tensor imaging and quantitative histological validation. NMR Biomed. 2014;27:1378–86.
Figures