4858
High Resolution T1-Perfusion using Compressed-SENSE for Glioma Grading1Department of Radiology and Imaging, Fortis Memorial Research Institute, Gurgaon, India, 2Philips Health Systems, Philips India Limited, Gurgaon, India, 3Center for Biomedical Engineering, Indian Institute of Technology Delhi, Delhi, India, 4Philips Innovation Campus, Philips India Limited, Bangalore, India, 5Department of Neurosurgery, Fortis Memorial Research Institute, Gurgaon, India, 6SRL Diagnostics, Fortis Memorial Research Institute, Gurgaon, India, 7Department of Neuroimaging & Interventional Radiology, National Institute of Mental Health and Neurosciences, Bangalore, India, 8Philips HealthTech Asia Pacific, Tokyo, Japan
Synopsis
T1-perfusion MRI derived relative cerebral blood volume (rCBV) is a key bio-marker for pre-surgical grading of gliomas; however, acquiring clinically relevant higher resolution T1-perfusion data with whole brain coverage is challenging due to possible loss in temporal resolution. This study takes the advantage of combining compressed-sensing with SENSE parallel-imaging i.e., Compressed-SENSE (CSENSE), to develop high-resolution whole brain T1-perfusion with improved temporality. The CSENSE enabled T1-perfusion derived rCBV values successfully differentiated high and low grade gliomas and matched with the histopathological grading. The rCBV cut-off value from CSENSE assisted T1-perfusion was similar to the routine T1-perfusion without-CSENSE of histopathology-matched gliomas.
Introduction:
T1-perfusion MRI is one of the noninvasive quantitative methods to estimate hemodynamic and kinetic parameters for diagnosis and management of human gliomas [1]. To develop a robust whole-brain T1-perfusion method for generating quantitative data with higher spatial and temporal resolution by overcoming current clinical limitations, methods like compressed sensing [2] for MR perfusion imaging has already been realized [3,4]. In this study, compressed sensing technology is combined with the SENSE parallel-imaging infrastructure, i.e., Compressed-SENSE (CSENSE), for high-resolution dynamic T1-perfusion data acquisition in human brain by exploiting the multi-element receiver coil sensitivity variation and sparsity constraining. This study also examines the efficacy of this quantitative-CSENSE method in deriving key quantitative parameter of T1-perfusion to differentiate tumor grades, i.e., relative cerebral blood volume (rCBV), by comparing the CSENSE accelerated perfusion results with histopathological grading of gliomas along with the rCBV cut-off value of histopathology-matched tumors previously acquired without CSENSE acceleration.Methods:
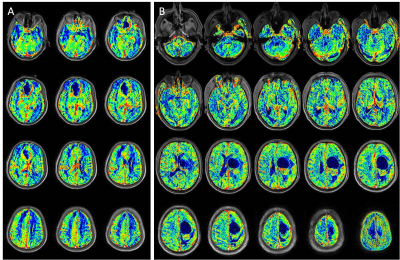
In this study, CSENSE enabled brain T1-perfusion data acquisition at 3.0 T (Ingenia, Philips, The Netherlands) included treatment naïve 26 patients; 19 with high grade and 7 with low grade gliomas. To quantify voxel-wise pre-contrast tissue longitudinal relaxation time T10 for computing leakage corrected CBV values [1], pre-contrast 2D T1-weighted TSE (TR/TE 360/10 ms), fast dual SE proton-density and T2-weighted fat suppressed images (TR/TE1/TE2 3500/23.2/90 ms) were acquired with 20 slices of thickness 6 mm, FOV= 230 × 230 mm2, reconstructed matrix= 256 x 256 and SENSE factor of 2. T1 dynamic perfusion images were acquired using a 3D T1-FFE with CSENSE factor 3.75. At the fourth time point of the dynamic data acquisition, 0.1 mmol/kg body weight of Gd-BOPTA (Multihance, Bracco, Italy) was administered intravenously at a rate of 3.0 ml/sec, followed by a 30-ml saline flush. A series of 640 images at 32 dynamics from 20 slices were acquired with a temporal resolution of 3.8 sec. T1 perfusion maps were generated using a home written script based on Leaky Tracer Kinetic Model of T1-perfusion [5,6]. Two experienced radiologists, blinded to the final histopathology, performed the ROI analysis by placing it on the slice showing the tumor with maximum rCBV and recorded the values. The rCBV values obtained from CSENSE-assisted T1-perfusion scan were compared with 26 histopathology-matched T1-perfusion data previously acquired without CSENSE and with 12 slices, FOV = 240X240 mm2 and matrix size = 128X128 with a temporal resolution of 3.95 s. All statistical analyses were performed with SPSS 16 software. Receiver operating characteristic (ROC) curve analysis was used to evaluate the performance of rCBV with and without CSENSE acceleration in discriminating between the high and low grades of tumors by comparing the area under the curve (AUC). A p value of <0.05 indicated a statistically significant difference.Results:
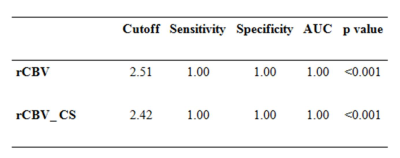
Compressed-SENSE accelerated T1-perfusion generated rCBV (Figure 1) values were able to successfully differentiate high grade gliomas from low grades with high sensitivity and specificity. The cut off between high grade and low-grade gliomas for high spatial resolution whole brain CSENSE enabled T1-perfusion is 2.42 and matched the non-CSENSE accelerated 12 slice data (Table-1, figure 1).Discussion:
T1-perfusion has the advantage over commonly used T2* based dynamic susceptibility contrast (DSC) perfusion as it provides both the quantitative hemodynamic and kinetic parameters of tumor, and it reliably generates rCBV values for the tumors in the presence of blood, calcifications, proximity to skull-base and in post-surgical lesions [7,8]. It is well known that perfusion derived cerebral blood volume correlates with tumor neoangiogenesis and hence the rCBV obtained from T1-perfusion MRI is considered one of the robust noninvasive methods for pre-operative tumor grading. However, acquiring higher resolution T1-perfusion data with whole brain coverage is challenging and may increase acquisition time at the cost of temporal resolution that is essential for quantification of CBV. This study takes the advantage of CSENSE acceleration to develop an imaging methodology so that higher resolution T1-perfusion data in terms of both spatial and temporal resolution with whole brain coverage can be acquired with higher temporal resolution. The rCBV obtained from CSENSE accelerated T1-perfusion data was accurate as it matched with histopathology grading to differentiate high and low grade gliomas in this study. Moreover, the rCBV cut-off to separate high grade from low grade gliomas was comparable with the cut-off obtained from histology-matched non-CSENSE accelerated T1-perfusion data.Conclusion:
This study demonstrates the possibility to obtain higher spatial and temporal resolution quantitative T1-perfusion data with whole brain coverage using CSENSE accelerated T1 dynamic scans; and the rCBV generated using this method may be used for pre-surgical evaluation of glioma.Acknowledgements
NoneReferences
- Gupta PK, Saini J, Sahoo P, Patir R, Ahlawat S, Beniwal M, Thennarasu K, Santosh V, Gupta RK. Role of Dynamic Contrast-Enhanced Perfusion Magnetic Resonance Imaging in Grading of Pediatric Brain Tumors on 3T. Pediatr Neurosurg. 2017; 52: 298-305.
- Michael Lustig, David Donoho, and John M. Pauly. Sparse MRI: The Application of Compressed Sensing for Rapid MR Imaging. Magnetic Resonance in Medicine. 2017; 58:1182–1195.
- David S. Smith, E. Brian Welch, Xia Li, Lori R. Arlinghaus, Mary E. Loveless, Tatsuki Koyama, John C. Gore and Thomas E. Yankeelov. Quantitative effects of using compressed sensing in dynamic contrast enhanced MRI. Phys Med Biol., 2011; 56(15): 4933–4946
- SoHyun Hana, Jeffrey L. Paulsenb, Gang Zhuc, Youngkyu Songd, SongI Chund, Gyunggoo, Chod, Ellen Ackerstaffe, Jason A. Koutchere, and HyungJoon Choa. Temporal/spatial resolution improvement of in vivo DCE-MRI with compressed sensing-optimized FLASH. Magn Reson Imaging, 2012; 30(6): 741–752
- Singh A, Singh A, Haris M, Rathore D, Purwar A, Sarma M, Bayu G, Husain N, Rathore RK, Gupta RK. Quantification of physiological and hemodynamic indices using T(1) dynamic contrast-enhanced MRI in intracranial mass lesions. J Magn Reson Imaging. 2007; 26: 871-880.
- P. Sahoo, Rathore RKS, Awasthi R, Roy B, Verma S,Rathore D, Behari S, Husain M, Husain N, Pandey CM, Mohakud S, Gupta RK, “Subcompartmentalization of extracellular extravascular space (EES) into permeability and leaky space with local arterial input function (AIF) results in improved discrimination between high- and low-grade glioma using dynamic contrast-enhanced (DCE) MRI.,” J. Magn. Reson. Imaging, 2013, vol. 38, no. 3, pp. 677–688.
- Essig M, Shiroishi MS, Nguyen TB, Saake M, Provenzale JM, Enterline D, Anzalone N, Dörfler A, Rovira A, Wintermark M, Law M. Perfusion MRI: the five most frequently asked technical questions. Am J Roentgenol. 2013; 200: 24-34.
- Artzi M, Blumenthal DT, Bokstein F, Nadav G, Liberman G, Aizenstein O, Ben Bashat D. Classification of tumor area using combined DCE and DSC MRI in patients with glioblastoma. Journal of Neuro-oncology 2015, 121: 349-57
Figures