James Ahad1, Wei-Ching Lo2, Jesse Hamilton2, Dominique Franson2, Yun Jiang3, and Nicole Seiberlich2
1Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 2Case Western Reserve University, Cleveland, OH, United States, 3Radiology, University Hospitals, Cleveland, OH, United States
Synopsis
Cardiac MRF is a unique MRF technique that requires a new dictionary for each acquisition to account for heart rate variability. Dictionary simulation presents a barrier for accessibility to cardiac MRF techniques due to the lack of tools at the scanner. To this end, Bloch simulation tools were developed on the Gadgetron platform to allow for online dictionary simulation and pattern matching reconstruction. Results between prior MATLAB implementations and Gadgetron implementation were in agreement. Future work in optimization may allow for data feedback from the scanner, reducing the need for additional scan time for accurate quantification of T1 and T2 maps.
Purpose
Cardiac MRF (cMRF)
(1, 2) is an MRF technique that enables simultaneous acquisition of T1,
T2, and M0 maps in the heart. cMRF is unique in that a
new MRF dictionary must be simulated after every scan in order to incorporate
subject- and scan-specific variations in heart rate. Gadgetron (3) is an
open-source modular reconstruction platform that allows complex reconstruction
algorithms to interface directly with MRI scanners. Previously, MRF has been
implemented in the Gadgetron for brain and prostate mapping (4). In those
studies, the MRF dictionary was pre-computed once and used to generate maps for
all subsequent acquisitions. In this study, Bloch equation simulation tools
were implemented in Gadgetron to allow for online dictionary simulation and
pattern matching at the scanner to enable Gadgetron to be employed for cMRF
reconstructions. Methods
Cardiac MRF data were
acquired on a 3T Skyra scanner (Siemens Healthineers, Erlangen, Germany) in the
short-axis view during a breathhold using previously described sequence
parameters (1, 2, 5). Dictionary generation and
tissue property map reconstructions were performed on the conventional
MATLAB-Mex C platform and as well as the Gadgetron platform. The Gadgetron
system specifications were: 8GB Nvidia GeForce GTX 1080 Graphics Card, 4-core
5.1GHz i7 7700k processor and 32 GB of 2400 MHz DDR4 memory. The cMRF
dictionary was simulated on the Gadgetron platform using the Bloch equations
with T1 values of [20:10:2000 2020:20:3000 3050:50:4000] and T2
values of [4:2:100 105:5:200 210:10:300 320:20:500] for a dictionary size of
22,866 entries. After L-2 normalization, the dictionary was compressed using
the SVD along the time dimension from 750 time points to 46 temporal basis
vectors (6). The k-space data were PCA coil compressed and SVD compressed
before NUFFT. For NUFFT, the MATLAB-Mex C reconstruction used the Fessler
toolbox (7), and the Gadgetron reconstruction used the Sorensen algorithm (8).
For the Gadgetron reconstruction, the resulting aliased images are adaptive
coil combined (9). Tissue property maps were generated by dot product matching
between the pixel timecourses and the dictionary (10). The root mean square
error (RMSE) between the cMRF dictionaries calculated in MATLAB and on the
Gadgetron were computed before SVD compression. In addition, the mean T1 and
T2 of the heart wall was obtained from manually drawn ROIs on the
parameter map outputs from MATLAB and Gadgetron.
Results
cMRF dictionary simulation time was 1220s
on MATLAB-Mex C and 2146s on Gadgetron. The RMSE between dictionaries was
7.6x10-6, indicating that the Bloch equation simulation on the
Gadgetron platform yields equivalent dictionary entries to MATLAB. The tissue
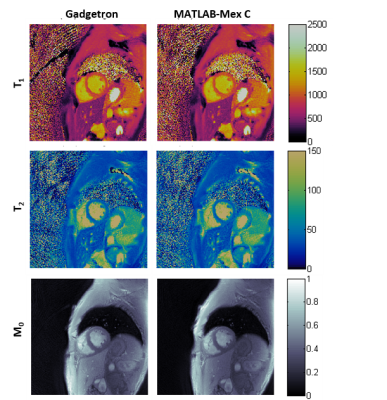
property maps show good agreement for T2 with slight differences in T1 values for the heart wall (Table 1). Figure 1 shows T1, T2 and M0 maps generated using both Gadgetron and MATLAB-Mex C
reconstructions.Discussion and Conclusion
Cardiac MRF requires the dictionary to be
simulated after data collection to account for heart rate variations, and thus
this step is essential for on-line tissue property mapping in the heart with
MRF; because clinical translation of MRF requires on-scanner reconstruction
without the cumbersome transfer of data to an off-line reconstruction engine,
this work opens the way for clinical adoption of cMRF. Tissue property maps
from both reconstruction platforms showed good agreement with each other, with
variation due to differences in pulse sequence modeling and the gridding
algorithm. While the Gadgetron-based MRF dictionary simulation had slower
performance, it competed against highly optimized MATLAB-Mex C code. While
MATLAB-Mex C code is efficient, it is narrow in application. In contrast, the
Gadgetron-based cMRF reconstruction can be employed at any modern MRI scanner.
The availability of such a vendor-independent cardiac MRF reconstruction system
will enable the evaluation of cMRF at other clinical sites. In addition, the
Gadgetron reconstruction has considerable space for further optimization,
particularly via GPU acceleration. With on-the-fly dictionary generation
capabilities, it may be possible to generate the MRF dictionary in real-time to
determine when enough data have been collected for accurate T1 and T2
quantification in order to reduce MRF data collection times.
Acknowledgements
R01HL094557, R01DK098503, R01EB016728, Siemens Medical Solutions.References
:
(1) Hamilton et al, MRM 2017. (2) Hamilton et al, JCMR 2016;18:W1. (3) Hansen
et al, MRM 2012. (4) Lo et al. Proc. of ISMRM Workshop on MRF 2017. (5) Hamilton
et al. Proc. Of ISMRM Workshop on MRF 2017. (6) McGivney et al. IEEE TMI 2014.
(7) Fessler et al, IEEE TSP 2003. (8) Sorensen et al. IEEE TMI 2008. (9)
Inati et al. Proc. of ISMRM. 2013, #2672. (10) Ma et al. Nature. 2013.