4781
Influence of contrast, sequence, threshold, and observer on 3D segmentation of the right ventricular outflow tract for intervention planning1Pediatric Heart Center, Cardiology, University Children's Hospital Zurich, Zurich, Switzerland, 2Pediatrics, University of Texas Southwestern Medical Center, Dallas, TX, United States, 3Division of Imaging Sciences and Biomedical Engineering, King's College London, London, United Kingdom
Synopsis
Three-dimensional reconstructions of cardiac magnetic resonance (CMR) datasets vary with contrast, image sequence, segmentation threshold, and manual post-processing. These influences need to be assessed before CMR-based models are used for interventional planning.
Three-dimensional segmentations of the right ventricular outflow tract (RVOT) from twelve patients with three different angiography sequences were compared between and within observers using different or the same thresholds.
Thresholding was sequence-dependent and did not significantly change object volumes. Minimal diameters of 3D reconstructed RVOTs showed clinically significant variation with different thresholds and between observers.
Interventional planning should rely on source images, not three-dimensional reconstructions, for quantitative information.
Introduction
Three-dimensional (3D) reconstructions of cardiac magnetic resonance (CMR) datasets are subject to variation based on the use of contrast, image sequence, threshold chosen by the operator, and manual postprocessing 1. If CMR-derived patient-specific objects are used for interventional or surgical planning, knowledge of the potential biases and post-processing errors is required to make informed decisions.Methods
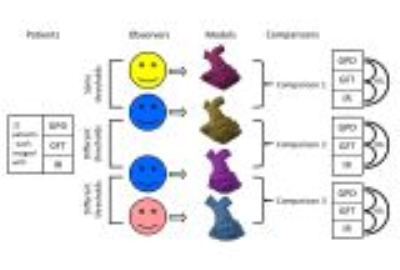
3D datasets from twelve patients with congenital heart disease who had contrast-enhanced CMR angiography (MRA) with gadopentetate dimeglumine (GPD) and with gadofosveset trisodium (GFT), and an inversion-recovery 3D whole heart sequence (IR), were segmented using Mimics version 19.0 software (Materialise Medical NV, Belgium). Objects were compared 1) between different observers using the same threshold, 2) between segmentations made by the same observer using different thresholds, and 3) between different observers using different signal thresholds (figure 1). The models were trimmed to leave only the right ventricle and main, right, and left pulmonary arteries. Models were aligned in 3-matic version 11.0, then borders were trimmed on both objects simultaneously, with several iterations. Subtractions and aggregate error objects were created using Boolean operations. Volumes of the models and circumferences of the right ventricular outflow tracts (RVOTs), relative overlap (similarity coefficients), and differences between objects were calculated.Results
The lower threshold limits were lower for IR than for MRA with GPD (202 ± 82 vs. 404 ± 136; p<0.001) and with GFT (398 ± 177; p<0.001). A constant threshold did not improve interobserver reliability of object volumes, with intraclass correlation coefficients ranging from 0.878 (IR; different observer, same threshold) to 0.99 (GFT; different observer, different threshold). Similarity scores between objects according to Dice and Sorenson were overall high (different observer, same threshold: 0.86-0.97; same observer, different threshold: 0.81-0.95; different observer and threshold: 0.9-0.97). No differences were found between sequences regarding the attainable similarity between observers. Minimal RVOT diameters differed on average by -0.6 to 1.1 mm between observers using the same threshold (widest limits of agreement (LOA): IR; -4.2 – 5.6 mm), by -1.9 to -1.3 mm between objects from the same observer using different thresholds (widest LOA: GPD; -7.7 to 5.1 mm), and by 0.4 to 1.4 mm between observers using different thresholds (widest LOA: GPD; -3.7 to 6.5 mm).Discussion
Thresholding was sequence-dependent and was a major determinant of object volumes. Differences in minimal diameters between 3D models were similar to reported reproducibilities of aortic diameters in two-dimensional measurements of MRA and 3D SSFP images 2, 3 and can be clinically relevant.Conclusion
Segmented objects show high reproducibility of volume and shape, but should not be relied upon without validation against source images when cardiovascular structures are segmented for interventional or surgical planning. Automatic segmentation tools should be further developed to minimize inter-operator errors.Acknowledgements
No acknowledgement found.References
1. Tandon A, Byrne N, Nieves Velasco Forte Mde L, et al., Use of a semi-automated cardiac segmentation tool improves reproducibility and speed of segmentation of contaminated right heart magnetic resonance angiography. Int J Cardiovasc Imaging 2016, 32 (8), 1273-1279.
2. Potthast S, Mitsumori L, Stanescu LA, et al., Measuring aortic diameter with different MR techniques: comparison of three-dimensional (3D) navigated steady-state free-precession (SSFP), 3D contrast-enhanced magnetic resonance angiography (CE-MRA), 2D T2 black blood, and 2D cine SSFP. J Magn Reson Imaging 2010, 31 (1), 177-184.
3. Veldhoen S, Behzadi C, Derlin T, et al., Exact monitoring of aortic diameters in Marfan patients without gadolinium contrast: intraindividual comparison of 2D SSFP imaging with 3D CE-MRA and echocardiography. Eur Radiol 2015, 25 (3), 872-882.