4779
Methodology for Non-Diseased Myocardium Part Segmentation of Delayed Enhancement 3D MRI by Using Watershed and Shape Priors1Le2i FRE2005, CNRS, University of Burgundy, Auxerre, France, 2Service de Spectroscopie-RMN, CHU de Dijon, Dijon, France
Synopsis
Previous approaches on left ventricle segmentation from DE-MRI have focused on the extraction of myocardium or just diseased region in short axis orientation. However these studies are not well suited for the segmentation of non-diseased myocardium region on DE-MRI. This paper presents a novel semi-automatic segmentation method of non-diseased myocardium region segmentation on diseased patient heart from DE-MRI based on watershed algorithm with shape priors application. To assess the results segmented images were compared with gold standard images performed by an experienced user. Novel solution for DE-MRI segmentation has been proposed, which brought promising results of measured parameters.
1. Introduction
Cardiac MR (CMR) provides information about structure, perfusion and contractile function of the heart. One of the MR techniques used to assess the viability of myocardium is DE-MRI1. To obtain quantitative results on the myocardium region, the application of segmentation method will be necessary. Previous approaches on left ventricle segmentation from DE-MRI have focused on the extraction of myocardium2-6 or just diseased region7-11. However these studies did not take into account the segmentation of non-diseased myocardium region from DE-MRI. The segmentation of non-diseased myocardium from DE-MRI, has some useful applications. For instance it can simplify the PET-MR registration process, because the strongest PET signal is expected on the areas of a healthy part of myocardium. Our method can be also used to differentiate the non-diseased and diseased part on cine-MRI. Several studies have been carried out on the segmentation of myocardium from cine-MRI by using the combination of shape priors and with usual segmentation method12,13. Shape priors significantly improves the results of myocardium segmentation on cine-MRI. The core problem of the left ventricle segmentation is the complex geometry of the myocardium, which is presented in various plane. However, the similarity between the MR signals on the myocardium and its surrounding region induces a low signal to noise ratio coefficient of DE-MRI. To deal with this problem, we propose a semi-automatic segmentation method for DE-CMR images dedicated to detect non-diseased myocardium part. Our method is based on watershed algorithm and shape priors application.2. Methods
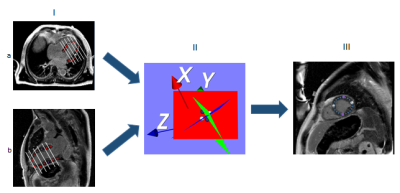
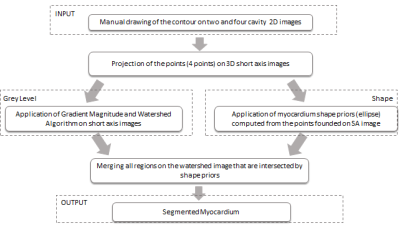
Vincent and Soilles presented a technique based on the watershed algorithm, which is applied to the gradient magnitude image data and produces the small volume primitives14, 15. The output of watershed segmentation is the image, which represents the homogeneous areas of gray level intensities, instead of every pixel representation. Watershed approach was selected in order to obtain the map of homogeneous areas on the image. Shape priors was applied to select the regions of pixels intensities which include non-diseased myocardium part. To obtain a left ventricle segmentation in 3D, user has to position the points on two and four cavity 2D DE-MR images to define the shape of the myocardium. The points are projected to 3D short axis images. For each short axis image we obtained four points which are used to define the shape of myocardium as an ellipse(see Fig.1). The gray level information is used as a reference value to choose the non-diseased parts of myocardium and excluded diseased regions, which were usually included in the applied shape (Fig.2).
Results
The segmentation process was performed on 160 short axis images. The images were acquired from 20 patients with different cardiac disorders (myocardial infraction, hypertrophic cardiomyopathy and normal heart). All patients underwent CMR in 1.5T magnet with late gadolinium enhancement (LGE) sequence in Centre Hospitalier Universitaire in Dijon.
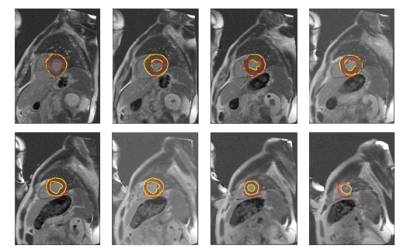
The quantitative analysis were performed to assess the difference between the gold standard performed by experienced user and segmented image obtained by using our method (see fig.2).
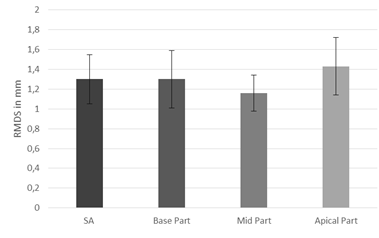
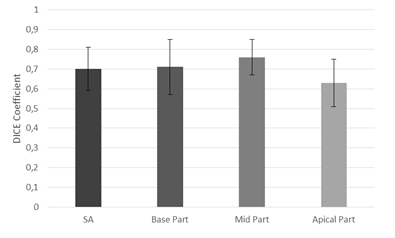
To assess the results the DC and RMSD were computed. The value of RMSD was equal 1.3±0.25mm and the value of DC was equal 0.7±0.11. The best value of these parameters was obtained at mid level (RMSD=1.16 ±0.18mm, DC = 0.76± 0.09), and base level (RMSD=1.3±0.29mm, DC=0.71±0.14) of myocardium. At apical level (RMSD=1.43±0.29mm, DC=0.63±0.12) the segmentation is less efficient due to the lower contrast between tissue in this part of myocardium (see fig.3 and 4).
Slightly low value of DC occurred in blurred and almost not visible edges between myocardium and tissues around. The value of RMDS is relatively low so it shows that the contour obtained by our method is close to the contour plotted by an expert.
Discussion and Conclusion
Cardiac MR Images are usually blurred with almost invisible boundary between lung and myocardium. Automatic detection of the regions of myocardium based on shape priors greatly simplifies the segmentation process. The segmentation of the images which represents different cardiac diseases, often brought good value of measured parameters. This method of non-diseased myocardium segmentation will be applied as one of the step in our 3D PET-MR registration method and to cine-MRI segmentation. Segmented images can be used to build 3D model of the non-diseased myocardium region. In conclusion, we have proposed a novel solution for non-diseased myocardium region segmentation from DE-MRI, which brought promising results.Acknowledgements
No acknowledgement found.References
- Kramer, Christopher M., et al. "Standardized cardiovascular magnetic resonance (CMR) protocols 2013 update." Journal of Cardiovascular Magnetic Resonance 15.1 (2013): 91.
- Chenoune, Y., et al. "Methodology for jointly assessing myocardial infarct extent and regional contraction in 3-D CMRI." IEEE Transactions on Biomedical Engineering 59.9 (2012): 2650-2659.
- Ciofolo, Cybele, et al. "Automatic myocardium segmentation in late-enhancement MRI." Biomedical Imaging: From Nano to Macro, 2008. ISBI 2008. 5th IEEE International Symposium on. IEEE, 2008.
- Dikici, Engin, et al. "Quantification of delayed enhancement MR images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Berlin, Heidelberg, 2004.
- Wei, Dong, et al. "Myocardial segmentation of late gadolinium enhanced MR images by propagation of contours from cine MR images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Berlin, Heidelberg, 2011.
- Xu, Robert S., et al. "Myocardial segmentation in late-enhancement MR images via registration and propagation of cine contours." Biomedical Imaging (ISBI), 2013 IEEE 10th International Symposium on. IEEE, 2013.
- Detsky, Jay S., et al. "Reproducible classification of infarct heterogeneity using fuzzy clustering on multicontrast delayed enhancement magnetic resonance images." IEEE transactions on medical imaging 28.10 (2009): 1606-1614.
- Hennemuth, Anja, et al. "A comprehensive approach to the analysis of contrast enhanced cardiac MR images." IEEE Transactions on Medical Imaging 27.11 (2008): 1592-1610.
- Karim, Rashed, et al. "A method to standardize quantification of left atrial scar from delayed-enhancement MR images." IEEE journal of translational engineering in health and medicine 2 (2014): 1-15.
- Lu, Yingli, et al. "Automated quantification of myocardial infarction using graph cuts on contrast delayed enhanced magnetic resonance images." Quantitative imaging in medicine and surgery 2.2 (2012): 81
- Tao, Qian, et al. "Automated segmentation of myocardial scar in late enhancement MRI using combined intensity and spatial information." Magnetic Resonance in Medicine 64.2 (2010): 586-594
- Mahapatra, Dwarikanath. "Cardiac image segmentation from cine cardiac MRI using graph cuts and shape priors." Journal of digital imaging 26.4 (2013): 721-730.
- Alessandrini, Martino, et al. "Using a geometric formulation of annular-like shape priors for constraining variational level-sets." Pattern Recognition Letters 32.9 (2011): 1240-1249.
- Grau, Vicente, et al. "Improved watershed transform for medical image segmentation using prior information." IEEE transactions on medical imaging 23.4 (2004): 447-458.
- Vincent, Luc, and Pierre Soille. "Watersheds in digital spaces: an efficient algorithm based on immersion simulations." IEEE Transactions on Pattern Analysis & Machine Intelligence 6 (1991): 583-598.
Figures