4778
CMRDiffTools: A Processing and Analysis Tool for Cardiac Diffusion MR images1Université de Lyon, UJM-Saint-Etienne, INSA, CNRS UMR 5520, INSERM U1206, CREATIS, F-42023, Saint Etienne, France, 2Department of Radiology, The University Medical Centre Utrecht, Utrecht, Netherlands, 3Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland, 4Centre for Clinical Magnetic Resonance Research, University of Oxford, Oxford, United Kingdom, 5Cardiovascular Research Centre, Royal Brompton Hospital, London, United Kingdom, 6Department of Radiological Sciences, University of California, Los Angeles, CA, United States
Synopsis
CMRDiffTools is an OsiriX/Horos plug-in for simplifying and streamlining the processing and analysis tasks of Cardiac Diffusion MR images. This tool includes state-of-the-art methods in image processing of diffusion weighted images, assembled in suitable workflows to be executed in a single pipeline. The CMRDiffTools plug-in empowers the user with interactive data exploration and visualization of specific cardiac fiber metrics such as helix, transverse and sheet angle.
Motivation and Objectives
The execution of image processing workflows to address specific issues of cardiac images (i.e. respiratory-motion induced image misalignment) relies on computer-based algorithms. Several biomedical image processing tools have been developed in order to provide a workspace for this task (i.e. 3D Slicer 1 and MITK Diffusion 2). However, these applications often provide a generic environment for the execution of image processing algorithms such as image filters for a wide range of biomedical image formats (MetaIO, NIfTI, imported DICOM, etc.). The generic aspect of such tools requires the user to perform additional steps (data import/export, format conversion, third-party software integration, etc.), which diverts attention from the main purpose, data processing and analysis. Accordingly, there is a need for a framework to establish a natural interaction between the user, Cardiac Diffusion MR (CDMR) data and cardiac-specific state-of-the-art workflows. Moreover, a common tool within the Cardiovascular Magnetic Resonance (CMR) research community would facilitate and foster reproducible research 3.
The objective of the present project was to develop an intuitive and effective research tool that can be deployed in clinical and academic settings. In addition, CMRDiffTools 4 aims to collect and make available state of the art computational methods developed by several international teams, for the benefit of the CMR research community.
Data Management and Workflow Execution
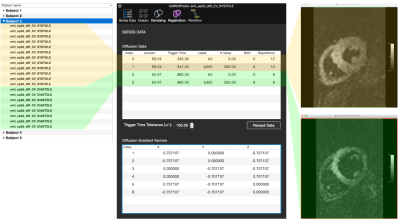
CMRDiffTools is implemented as a plug-in for the OsiriX DICOM viewer 5, a well-known platform for navigation and visualisation of multidimensional DICOM images. OsiriX can be deployed in clinical infrastructures connected to PACS and MR scanners streamlining data storage (DICOM standard), processing and analysis tools. The user can work directly on the DICOM study/series of interest using advanced processing and analysis methods. The CMRDiffTools plug-in handles the complexity of the input data (Figure 1) displaying an overview grouping datasets for reconstruction and then providing details on demand.
CMRDiffTools contains 5 modules: Pre-Processing Tools, 2D Tensor-Based Analysis, Multi-exponential Analysis, 3D Fiber-Based Analysis and Documentation. This structure establishes a simple and natural dialogue between the workflows (pre-processing data, analysing tensor data, etc.) and capabilities of the tool (image registration, tensor reconstruction, helix angle calculation, etc.).
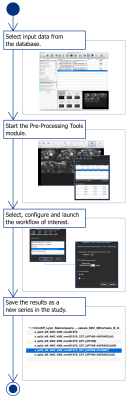
Each module contains sections; each of these sections has a specialized purpose. For instance the Pre-Processing Tools module has the sections Registration and Denoising, the user can execute one of registration/de-noising methods independently, or launch a workflow, a sequence of available methods in the module (i.e. a rigid registration followed by an average of repetitions). Figure 2 shows the sequence of actions required to launch a pre-processing workflow.
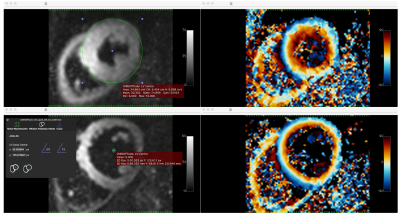
Once a pre-processing workflow is complete, the user can open another module (i.e. 2D Tensor-Based Analysis) to perform a different type of workflow. Figure 3 shows the results of a tensor-based workflow (on data presented in figure 1) comprising of: average repetitions, tensor reconstruction and helix angle calculation performed in a single pipeline. The most time-consuming operation for the user is to define the centre (a point or Epicardial contour) of the left ventricular cavity on the current image view.
Evaluation and Results
The objective of the CMRDiffTools project is to make available state-of-the-art computational methods developed by CMR research community; accordingly, the research scientist (the CMRDiffTools user) is able to use a well-known pre-processing workflow (i.e. image registration, PCA signal enhancement and finally, a temporal maximum intensity projection filter 6) in a few steps (mouse clicks). This evaluation is intended to provide evidence on the benefits of a diffusion MR, cardiac-specific research tool by performing a common task in the field: the helix angle calculation after a data refinement process.
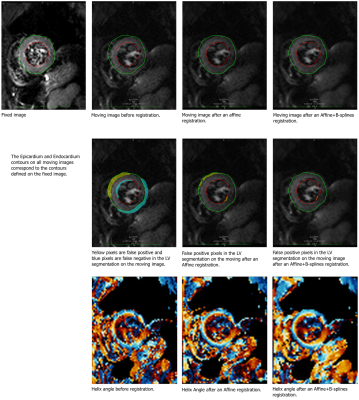
Figure 4 illustrates different pre-processing workflows (registration + average repetitions) followed by helix angle calculations. To highlight the differences between pre-processing workflow executions, pixels are classified by an evaluated segmentation of the Left Ventricle (LV): false positive if the pixel is part of the segmentation but not in the LV and false negative if the pixel is part of the LV but is not in the segmentation. This classification illustrates how large or small is the adjustment performed (by an affine transformation for instance) and the impact on the helix angle map. This visual assessment can be easily performed on the OsiriX/Horos platform with the CMRDiffTools plug-in.
Conclusion
CMRDiffTools is an OsiriX/Horos
plug-in to analyze Cardiac Diffusion MR images. Consequently, this
computer-based tool can be deployed in both clinical and research environments,
providing a bridge from state-of-the-art image processing methods
(contributions from the CMR scientific community) to its direct utilization in
a clinical environment. The infrastructure of OsiriX allows users to easily add additional computational methods developed
by the community.
Acknowledgements
No acknowledgement found.References
1. Pieper, S., Lorensen, B., Schroeder, W., and Kikinis, R. (2006). The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the medical image computing community, in 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro, 2006 (Arlington, VA: IEEE), 698–701. doi: 10.1109/ISBI.2006.1625012
2. Fritzsche, K., Neher, P., Reicht, I., van Bruggen, T., Goch, C., Reisert, M., et al. (2012). MITK diffusion imaging. Methods Inform. Med. 51, 441. doi: 10.3414/ME11-02-0031
3. Donoho, D. L. (2010) An invitation to reproducible computational research, Biostatistics. July; 11 (3): 385-388.
4. CMRDifftools website, https://www.creatis.insa-lyon.fr/osirix-dev/CMRDiffTools.html
5. Rosset, A. Spadola, L. and Ratib, O. (2004). OsiriX: An Open-Source Software for Navigating in Multidimensional DICOM Images, Journal of Digital Imaging. Sep;17(3):205-216.
6. Wei H, Viallon M, Delattre BM, et al. In vivo cardiac diffusion tensor imaging in free-breathing conditions. Journal of Cardiovascular Magnetic Resonance. 2013;15(Suppl 1):P231. doi:10.1186/1532-429X-15-S1-P231.
Figures