4773
Towards automatic analysis of 4D Flow MRI: Automatic cardiac segmentation1Department of Medical and Health Sciences, Division of Cardiovascular Medicine, Linköping University, Linköping, Sweden, 2Center for Medical Image Science and Visualization (CMIV), Linköping University, Linköping, Sweden, 3Sectra AB, Linköping, Sweden, 4Department of Clinical Physiology, Department of Medical and Health Sciences, Linköping University, Linköping, Sweden
Synopsis
One of the most important post-processing steps in the analysis of cardiac MR images is the segmentation of the blood pool, usually relying on manual delineation of the cardiac anatomy by an expert observer. Obtaining high quality segmentations of 4D Flow MR images acquired without the use of blood pool agents is especially challenging due to the low contrast between the blood and the myocardium present in these images. We propose an automatic multi-atlas segmentation technique that generates four-dimensional segmentations of the cardiac chambers and great thoracic vessels in 4D Flow MR images.
Introduction
4D Flow MRI is a time-resolved three-dimensional (4D) phase-contrast MRI technique that includes three-directional velocity encoding1. Typically, 4D Flow MRI acquisitions encompass the entire heart and the major thoracic vessels in one volume. In order to obtain useful segmentations of the heart chambers, semi-automatic segmentation methods are commonly applied on balanced Steady State Free Precession (bSSFP) MR images, superimposing the resulting labels over the 4D Flow MRI. However, bSSFP images suffer from a number of problems such as being prone to displacement errors caused by respiratory motion and large slice thickness2,3. We propose a multi-atlas segmentation method, which uses only 4D Flow MRI data, to automatically generate four-dimensional segmentations of the cardiac chambers and major thoracic vessels.Methods
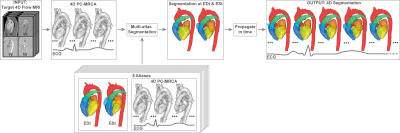
Atlas-guided segmentation uses the information contained in a previously segmented image, known as atlas, in conjunction with image registration to generate the desired segmentation4. Multi-atlas segmentation uses multiple atlases to capture the inter-subject variability of the anatomy to be segmented5,6.
For this work, we manually segmented eight datasets at two timeframes, viz. end-systole and end-diastole, to serve as atlases. Subsequently, the steps to generate a four-dimensional cardiac segmentation were as follows:
- A 4D Phase-Contrast MR CardioAngiography (4D PC-MRCA)7, was generated for each of the atlases and for the input dataset to be segmented.
- The PC-MRCAs of each atlas and the input dataset were non-rigidly registered in order to propagate the labels on each atlas to the input dataset. This step was done for both timeframes included in each atlas.
- The labels corresponding to all the atlases were combined into one final segmentation using the STAPLE algorithm8. The result of this step was a segmentation for the input dataset at two timeframes: end-systole and end-diastole.
- Non-rigid registration was applied between the timeframes of the target dataset, generating transformations that were applied to the previously obtained segmentation. This resulted in a four-dimensional segmentation of the regions labeled in the atlases.
A flowchart of the proposed technique can be seen in Figure 1.
Evaluation of the proposed method was performed on a group of 110 subjects: 27 healthy volunteers, and 83 patients with suspected or diagnosed chronic ischemic heart disease. 4D Flow MRI datasets were acquired using a 3T Philips Ingenia scanner with a free-breathing, navigator gated sequence. Scan parameters included: Candy cane view adjusted to cover both ventricles, VENC 120-150cm/s, flip angle 10°, echo time 2.5-2.6ms, repetition time 4.2-4.4ms, spatial resolution 2.7x2.7x2.7mm, and elliptical k-space acquisition. For evaluation purposes, cine MR images were acquired using a bSSFP protocol. This resulted in a short-axis stack with 1x1mm resolution, and slice thickness of 8mm. Manual segmentations were performed on these images to compare with the automatic segmentations generated by the proposed method.
Results
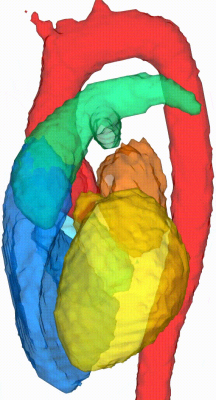
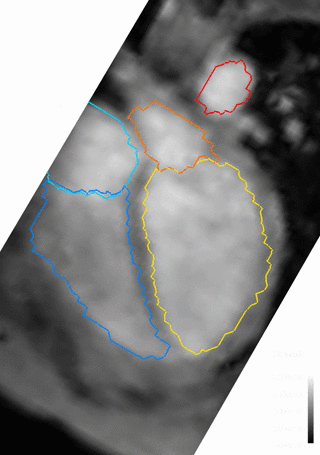
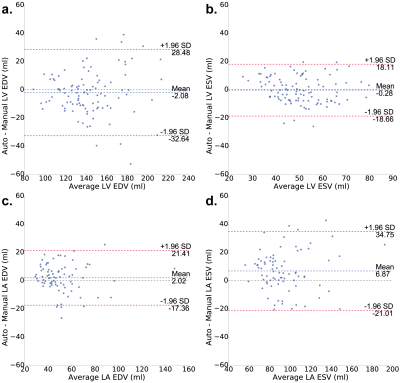
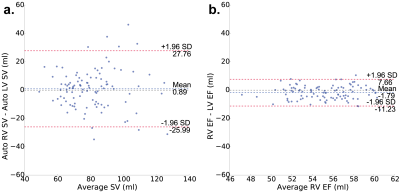
An example of the four-dimensional segmentations obtained can be seen in Figures 2 and 3, which include the cardiac chambers and major thoracic vessels. Figure 4 shows Bland-Altman plots comparing the left ventricular (LV) and left atrial (LA) volumes obtained through manual segmentation of the bSSFP images with the volumes obtained automatically. Stroke volumes (SV) of the LV and right ventricle (RV) were also compared, and are shown in Figure 5.Discussion
The proposed method allows for automatic generation of four-dimensional segmentations of the cardiac chambers and major vessels in 4D Flow MRI. Evaluation on 110 datasets from healthy volunteers and patients resulted in good agreement between manual and automatic segmentations. The quality of the results obtained were comparable to automatic segmentation methods attempted previously on bSSFP images9–12. Automation of the segmentation process can contribute in the assessment of 4D Flow MRI, particularly on large scale data, by making the process faster, more reliable, and repeatable. 4D Flow MR images typically have lower resolution when compared to the in-plane resolution of the bSSFP MR images, which affects the sharpness of the myocardium and can hinder the visualization of small structures. This limitation is also present in the segmentations generated by the proposed technique. The implementation evaluated in this study took approximately 25 minutes to generate a segmentation for each dataset using parallel computing on a 3.5GHz, 6-core CPU with 64GB RAM. However, further development using specialized hardware can result in significantly reduced computational times.Conclusion
Multi-atlas segmentation of 4D Flow MRI datasets permits automated and accurate segmentation of the cardiac ventricles and the large thoracic vessels. The resulting segmentations are four-dimensional and can facilitate flow assessment in the entire thoracic cardiovascular system, thus increasing the feasibility of 4D Flow MRI in the clinical setting.Acknowledgements
This work was partially funded by the FP7-funded project DOPPLER-CIP [grant number 223615]; the European Union's Seventh Framework Programme (FP7/2007-2013) [grant number 310612]; the Swedish Research Council [grant number 621-2014-6191]; and the Swedish Heart and Lung Foundation [grant number 20140398].
The authors have no relevant conflicts of interest to disclose.
References
1. Markl M, Kilner PJ, Ebbers T. Comprehensive 4D velocity mapping of the heart and great vessels by cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2011;13(1):7.
2. Holland AE, Goldfarb JW, Edelman RR. Diaphragmatic and cardiac motion during suspended breathing: preliminary experience and implications for breath-hold MR imaging. Radiology. 1998;209(2):483-489.
3. Scott AD, Keegan J, Firmin DN. Motion in cardiovascular MR imaging. Radiology. 2009;250(2):331-351.
4. Rohlfing T, Brandt R, Menzel R, Russakoff D, Maurer Jr. C. Quo Vadis, Atlas-Based Segmentation? In: Suri J, Wilson D, Laxminarayan S, eds. Handbook of Biomedical Image Analysis. Springer US; 2005:435-486.
5. Rohlfing T, Brandt R, Menzel R, Maurer CR. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004;21(4):1428-1442.
6. Doan NT, Orban de Xivry J, Macq B. Effect of inter-subject variation on the accuracy of atlas-based segmentation applied to human brain structures. In: SPIE Vol 7623. ; 2010:76231S-76231S-10.
7. Bustamante M, Gupta V, Carlhall C-J, Ebbers T. Improving visualization of 4D flow cardiovascular magnetic resonance with four-dimensional angiographic data: generation of a 4D phase-contrast magnetic resonance CardioAngiography (4D PC-MRCA). J Cardiovasc Magn Reson. 2017;19(1):47.
8. Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23(7):903-921.
9. Jolly M-P, Guetter C, Lu X, Xue H, Guehring J. Automatic Segmentation of the Myocardium in Cine MR Images Using Deformable Registration BT - Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges: Second International Workshop, STACOM 2011, Held in Conjunction wit. In: Camara O, Konukoglu E, Pop M, Rhode K, Sermesant M, Young A, eds. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012:98-108.
10. Bai W, Shi W, O’Regan DP, et al. A probabilistic patch-based label fusion model for multi-atlas segmentation with registration refinement: Application to cardiac MR images. IEEE Trans Med Imaging. 2013;32(7):1302-1315.
11. Queiros S, Barbosa D, Heyde B, et al. Fast automatic myocardial segmentation in 4D cine CMR datasets. Med Image Anal. 2014;18(7):1115-1131.
12. Shahzad R, Tao Q, Dzyubachyk O, Staring M, Lelieveldt BPF, van der Geest RJ. Fully-automatic left ventricular segmentation from long-axis cardiac cine MR scans. Med Image Anal. 2017;39:44-55.
Figures