4590
Across vendor comparison of multi-echo GRE R2* measurements in the kidney and a tissue-mimicking phantom1Radiology, UZ Brussel, Brussel, Belgium
Synopsis
BOLD MRI may serve as an indicator of blood oxygenation in the kidney and could potentially be used as a MRI biomarker in several kidney diseases. To be effective as a quantitative imaging biomarker, BOLD MRI must be an objective measure, independent of the MRI scanner model and MRI sequence implementation details. We have compared BOLD imaging across vendors in a tissue-mimicking phantom and in the healthy kidney by assessing R2* values in 12 concentric layers.
Introduction
BOLD MRI may serve as an indicator of blood oxygenation in the kidney [1], [2] and could potentially be used as a MRI biomarker in several kidney diseases. To be effective as a quantitative imaging biomarker, BOLD MRI must be an objective measure, independent of the MRI scanner model and sequence implementation details. In this study, we have compared BOLD imaging across vendors in a tissue-mimicking phantom and in the healthy kidney by assessing R2* values in 12 concentric layers. R2* is measured using a multi-echo gradient echo sequence on three MRI scanners from different vendors, while harmonizing acquisition parameters between vendors.Methods
An agar phantom was created according to the established FBIRN phantom recipe [3], which is well-suited for quality assurance of BOLD imaging in the brain. The agar gel was poured into a square plastic container of 175x120x48mm, with a volume of 0.65 liter. The phantom and a healthy volunteer with no history of kidney disease were scanned on three scanners in the same hospital, from vendors A, B, and C with a 70 cm bore, using a vendor-specific implementation of a multi-echo gradient-echo (GRE) sequence. The sequence parameters were based on the protocol used in [4]. After setting up the protocols on each scanner, the common denominator across all three vendors was determined. FOV 400mm, matrix 256x256, one 5mm coronal slice. TR=76 ms (78 ms on vendor C), TE=6 to 54.4ms in steps of 4.4ms. Bandwidth was 270 Hz/pixel, or 35.71 kHz, or a fat-water shift of 1.6 ppm dependent on the vendor definition. Images were acquired during breath hold, using a 36-channel body coil with dielectric pad on vendor A, a 32-channel body coil on vendor B and an 18-channel body coil on vendor C. Measurements were repeated three times.
All images were converted to Nifti using mrconvert from the MRtrix3 package [4]. Background noise standard deviation was calculated as $$$\sigma=\sqrt(\eta/2L)$$$, with $$$\eta$$$ the mean noise signal in the kidney/phantom over all TEs, and L the number of receive coil channels [5]. Parameters for NLM filtering: search window 11x11, similarity window 5x5, h=1.5$$$\sigma$$$ [5]. R2* estimation using non-linear Levenberg-Marquardt estimation of the squared signal to a single exponential model [5]. Left and right kidney were segmented in FSLview. R2* curves were created by taking the mean R2* across 12 layers in the kidney or a square ROI in the phantom using the 12-layer concentric object method [1], [3], implemented in Matlab. 95% confidence intervals (CI) were calculated as $$$1.96*\sigma/\sqrt(n)$$$, with the R2* standard deviation and n the number of samples in each concentric layer.
Results
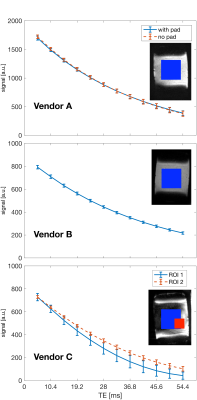
Fig 1 shows the signal decay versus echo time in a square ROI in the phantom. On vendors A and B, the decay is close to exponential, while in vendor C the curves deviate from an exponential curve. On closer inspection, the images from vendor C show large regions of signal loss at higher echo times compared to vendors A and B. A second, smaller, ROI was drawn on the last-echo image in a region containing signal, and is showing signal decay closer to exponential decay.
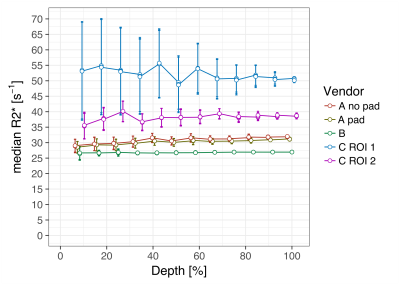
In fig 2, the R2* relaxation rate is shown as function of the depth. Mean R2* [s-1] over all the layers is 28.5 for vendor A with dielectric pad, 30.8 without using the pad, 26.8 for vendor B, 52.1 for vendor C and 38.2 for vendor C in ROI 2. R2* standard deviations for vendor C are considerably larger than in vendors A and B.
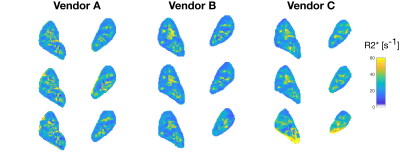
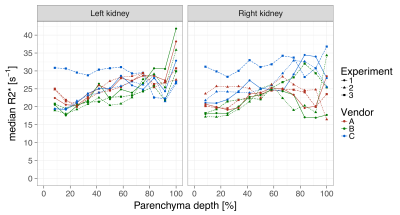
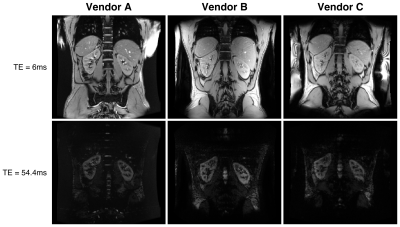
In the volunteer, R2* maps across three vendors are visually similar as shown in fig 3. The concentric layer method shows that R2* values between the three scanners are in the same range (Fig 4), but deeper layers show large variations. R2* values for the left and right kidney are similar for all vendors, while for vendor C the values are higher in one of the experiments. GRE images, Fig 5, at TE=6 and TE=54.4 ms show comparable image quality for vendors A and B, but show artefacts and signal loss in vendor C.
Discussion/Conclusion
The three vendors each have a different implementation of a multi-echo gradient echo sequence, but kidney R2* values are in the same range across vendors in this pilot study on a single volunteer. In the tissue-mimicking phantom, R2* values are comparable between vendors A and B, but vendor C shows a large deviation compared to the other vendors. This initial experiment will serve as a starting point for harmonization of BOLD measurements in the kidney.Acknowledgements
COST Action CA16103. The authors thank Dr. Bastien Milani and Dr. Matthias Stuber (CHUV, Lausanne, CH) for providing the full scan protocol from reference [4].References
[1] Front. Physiol., vol. 7, pp. 1–7, 2017. [2] Acta Physiol., vol. 213, pp. 19–38, 2015. [3] JMRI 23, no. 6 (June 2006): 827–39. doi:10.1002/jmri.20583.[4] Magn. Reson Imaging, vol. 33, no. 3, pp. 253–261, 2015. [4] https://github.com/MRtrix3/mrtrix3, 2012. [5] Magn. Reson Med., vol. 72, pp. 260–268, 2014.Figures