4578
Fast, automated slice prescription of standard anatomical planes for fetal brain MRI1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States, 4Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 5Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 6German Center for Neurodegenerative Diseases, Bonn, Germany
Synopsis
Motion limits MRI of the fetal brain to rapid 2D acquisitions with low resolution. Even with such sequences, it is challenging to obtain images aligned with standard anatomical planes of the brain, and views rendered retrospectively across slices typically suffer from artifacts due to between-slice motion. Here, we present an automated on-scanner slice prescription: immediately before sequence execution, the FOV/slice tilt is updated to match the orientation and position of the brain, derived from the previous acquisition. The fast update is achieved by registration to a template.
Introduction
Fetal head motion and maternal respiration limit MRI of the fetal brain to rapid 2D sequences with low resolution, such as HASTE, that effectively freeze motion during the acquisition of individual slices. However, motion can occur between manual slice positioning and image acquisition, resulting in images with incorrect anatomical alignment. Typically, repeated acquisitions with manually corrected slice orientation are necessary to acquire images with the desired anatomical coordinates (sagittal/axial/coronal). Reslicing datasets is not an option, as image quality of views rendered across slices is dramatically reduced by between-slice motion and spin-history effects. These factors result in an inefficient use of scan time due to discarded acquisitions.
We present automated slice prescription of standard anatomical planes for fetal brain imaging, and assess the suitability of HASTE and FSL/FLIRT1 for unsupervised on-scanner orientation estimation. The orientation of the brain is derived from an initial scout HASTE acquisition by registration to an atlas. The next HASTE scan is automatically prescribed based on the relative rotations of the scout to the atlas.
Methods
The pipeline was fully implemented on a 3 T Prisma scanner (Siemens Healthineers, Erlangen, Germany) and runs in real time without the need for any additional equipment.
Anatomical reference and imaging
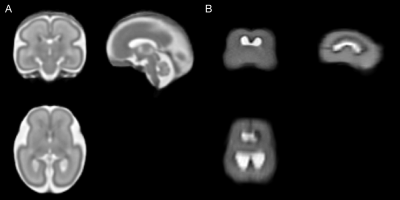
A fetal brain atlas at 23-37 weeks gestational age (GA)2 was used as anatomical reference (see Figure 1A). Images were acquired with HASTE (TR/TE 1600/100 ms, voxel size 1.3x1.3 mm2, matrix 256x256, 48 3-mm slices, BW 680 Hz/px).
Registration
The scout is cropped to a cubic box with side length of the 95th percentile of occipito-frontal diameter3, assuming the brain is approximately centered within the FOV. Immediately after acquisition the volume is written to the scanner’s file system using ITK4: to estimate any shift between the brain and the center of the FOV (instead of the isocenter), we modify the image metadata such that the image is centered at the origin.
To compensate for spin-history effects, we normalize slice intensities. We perform rigid-body registration with FLIRT based on correlation of ratio images. Non-brain voxels of the template are ignored. As the fetal brain can have any orientation in the scanner, the search range is extended to 360° and search intervals are halved.
Updates to the FOV
The FOV/slice tilt of the next scan is automatically aligned with the brain by rotating the known anatomical axes of the template based on the estimated transform. From these axes, the slice normal vector is selected according to a predefined anatomical plane. To avoid wrap-around, phase-encoding and readout direction are chosen by maximizing the angle between the phase and head-foot direction.
Results
On-scanner alignment
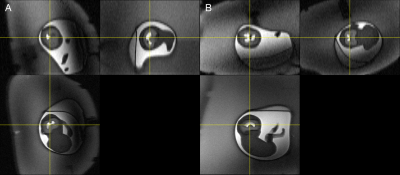
Automatic on-scanner FOV alignment was tested on a gravid abdomen phantom5 for initially oblique scan planes relative to the phantom fetal brain. The pipeline successfully reoriented the FOV with specified anatomical planes; a HASTE image of the phantom with masked non-brain voxels was manually aligned with the scanner axes and used as registration target (see Figure 1B). Images before/after automatic reorientation are shown in Figure 2.
Offline registration evaluation
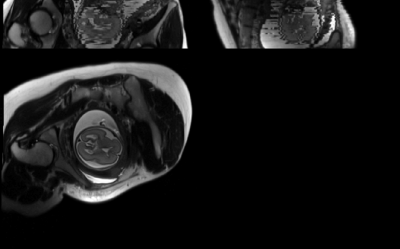
Registration was assessed offline for n=9 acquisitions from three fetuses (25/28/37 weeks GA), with varying slice orientation and motion. A typical dataset is shown in Figure 3. Performance was evaluated by visual inspection. Despite pronounced motion artifacts, FLIRT correctly aligned 67% of datasets, with complete misregistration of the remaining images. Factors in favor of successful alignment included high SNR and low GA, i.e. more signal from bright amniotic fluid/subarachnoid spaces. Figure 4 shows a typical image before/after registration.
Discussion
We demonstrated automatic slice prescription of standard anatomical planes for fetal brain MRI. The ~15-sec execution time would allow motion to occur between the scout and actual scan, but could be shortened by initializing FLIRT with previous transforms while reducing the search range. Further work may integrate our approach with motion correction, e.g. using vNavs6: these ~300-ms low-resolution navigators permits the estimation of motion during the scan.
Offline experiments indicated that further work is needed to increase registration robustness: between-slice motion violates the assumption of a consistent volume. As registration of neighboring slices would increase processing time, a more rapid acquisition such as EPI could reduce motion artifacts in the scout, and improve registration performance. It may also be useful to automatically locate the fetal brain from an initial abdominal scout7.
Conclusion
We present an automated on-scanner image plane prescription for fetal brain imaging. This pipeline addresses the need for manually determining the orientation of the moving brain from oblique views, which is challenging and time-consuming.Acknowledgements
This research was supported by grants NINDS U01AG052564, NIBIB R01EB008547, NICHD R01HD085813 and 4R00HD074649.References
1. Jenkinson M, Smith SM. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2):143-156.
2. Serag A, Aljabar P, Ball G, et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. NeuroImage. 2012;59(3):2255-65.
3. Snijders RJM, Nicolaides KH. Fetal biometry at 14-40 weeks’ gestation. Ultrasound Obstet. Gynecol. 1994;4(1):34-48.
4. Yoo TS, Ackerman MJ, Lorensen WE, et al. Insight Segmentation and Registration Toolkit (ITK). National Library of Medicine. https://itk.org. Accessed November 1, 2017.
5. Garcia-Polo P, Gagoski B, Guerin B, et al. An anthropomorphic MR phantom of the gravid abdomen including the uterus, placenta, fetus and fetal brain. Proc. ISMRM, Toronto, 2015.
6. Tisdall MD, Hess AT, Reuter M, et al. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magn. Reson. Med. 2012;68(2):389-399.
7. Keraudren K, Kuklisova-Murgasova M, Kyriakopoulou V, et al. Automated fetal brain segmentation from 2D MRI slices for motion correction. NeuroImage. 2014;101(1):633-643.
Figures