4562
Prediction of invasive placenta with quantitative placental textures features from clinical MRI scanHuaiqiang Sun1, Haoyang Xing1,2, Haibo Qu3, Yi Liao3, Xiaoxia Zhou4, Zhiyi Zhou4, Qiyong Gong1, and Shu Zhou4
1Huaxi MR Research Center, Department of Radiology, West China Hospital, Sichuan University, Chengdu, China, 2College of Physical Science and Technology, Sichuan University, chengdu, China, 3Department of Radiology, West China Second University Hospital, Sichuan University, Chengdu, China, 4Department of Obstetrics and Gynecology, West China Second University Hospital, Sichuan University, Chengdu, China
Synopsis
A machine learning framework that can predict invasive placenta with MRI texture features and identify significant relevant image features
Purpose:
Placenta accrete, which is defined as direct attachment of the placental trophoblast to the uterine myometrium, represents one of the most morbid conditions in modern obstetrics, with high rates of catastrophic perinatal hemorrhage, hysterectomy and intensive care unit admission. Because of the increased risk to the patient at the time of delivery, early and accurate diagnosis of abnormal placenta attachment can be of great utility to surgeons for preoperative planning and patient counseling. MRI is indicated in the diagnostic workup when the ultrasound evaluation is equivocal or for patients with high clinical risk factors for placenta accreta. MRI signs of abnormal placentation include marked placental heterogeneity, irregular thick intraplacental T2 dark bands and extensive intraplacental vascularity(1). However, characterization of placental heterogeneity and presence of abnormal T2 dark bands appeared to be subjective. Currently, there is no official standardization of MRI protocols and there are no large series addressing the interobserver variability for the evaluation of invasive placenta. The use of quantitative features extracted from medical image has been an attractive field of research to overcome the subjectivity of visual interpretation(2). The current study aim to find image signatures from texture analysis that can quantitatively represent MRI criteria for the diagnosis of abnormal placentation and to evaluate the performance of classifier constructed on identified image biomarkers.Methods:
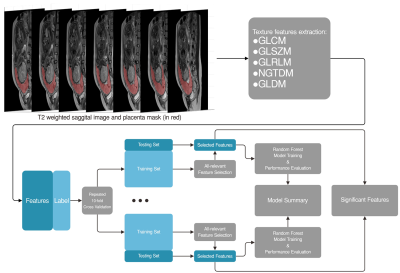
This retrospective study included 167 pregnancies (age from 25 to 35 years, mean 30.2 years) with placenta previa and suspicion of placenta accreta who underwent placenta MR imaging (gestational week at MRI scan from 27 to 35 weeks, mean 31.5 weeks) before delivery. T2 weighted images were acquired on a clinical 1.5T scanner with surface phase array coil using half-Fourier single-shot turbo spin echo imaging (TR=465ms, TE=80ms) sequence with 384*384 matrix over a field of view of 375*375 mm and 38 sagittal slices of 7 mm thickness. Ninety-nine pregnancies were surgical confirmed have placenta accreta at delivery and rest (68 pregnancies) had no invasive placenta. Placenta were manually delineation on 7 continues sagittal slices, which cover the suspect invasive site, of T2 weighted images. Texture features included the gray-level co-cocurrence matrix (GLCM) features (26 features), gray level size zone matrix (GLSZM) features (16 features), gray level run length matrix (GLRLM) features (16 features), neighboring gray tone difference matrix (NGTDM) features (5 features), gray level dependence matrix (GLDM) Features (15 features) were extracted from predefined placenta mask. The extracted features were input to all-relevant feature selection algorithm (implemented in R package “Boruta”), and random forest classifier was constructed based on selected features. To avoid selection bias, the feature selection and model construction procedure were embedded in repeated 10-fold cross-validation loops (Figure 1). The balanced accuracy, sensitivity/specificity, kappa scores were used to characterize the performance of the classifier. Texture features that were selected in all loops were identified as significant relevant features.Results:
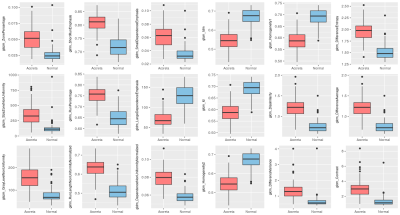
A total of 78 texture features were extracted from T2-weighted placenta image of each participant. In building and evaluation the classifier for discriminating normal and invasive placentation, 100 runs of 10-fold stratified cross-validation were performed, and the average performance of classifiers were computed from a total of 1000 folds. The mean balanced classification accuracy and kappa value achieved by our method via a repeated 10-fold cross-validation is 83.4%±7.2% and 0.67±0.14, using features selected by all-relevant feature selection algorithm. The mean sensitivity and specificity for placenta accreta is 89.4%±8.4% and 77.4%±14%, respectively. As we embedded the feature selection in cross-validation procedure, thus a total of 1000 feature subsets were obtained from each run of classifier construction. Eighteen significant relevant texture features, which were selected in all runs, are listed as: GLSZM_ZonePercentage, GLSZM_SizeZoneNonUniformity, GLSZM_GrayLevelNonUniformity, GLRLM_ShortRunEmphasis, GLRLM_RunPercentage, GLRLM_RunLengthNonUniformityNormalized, GLDM_SmallDependenceEmphasis, GLDM_LargeDependenceEmphasis, GLDM_DependenceNonUniformityNormalized, GLCM_Idm, GLCM_Id, GLCM_Homogeneity2, GLCM_Homogeneity1, GLCM_Dissimilarity, GLCM_DifferenceVariance, GLCM_DifferenceEntropy, GLCM_DifferenceAverage, GLCM_Contrast. The distribution of significant relevant features in two groups are shown in Figure 2Discussion and Conclusion:
It has been proved that MRI can be a reliable and reproducible tool for the detection of suspected placental invasion, but the diagnostic value significantly depends on observers’ experience(3). The current study established a quantitative analysis framework for invasive placenta prediction based on placental texture features extracted from routine clinical MRI images. The proposed framework overcome the subjectivity of visual interpretation and thus provide a useful objective adjunct to clinical evaluation for the risk of placenta accreta. The texture features identified by feature selection step also confirmed the signal heterogeneity within placenta has significant diagnostic value for predicting invasive placenta.Acknowledgements
No acknowledgement found.References
1. Lax A, Prince MR, Mennitt KW, Schwebach JR, Budorick NE: The value of specific MRI features in the evaluation of suspected placental invasion. Magn. Reson. Imaging 2007; 25:87–932.
2. Gillies RJ, Kinahan PE, Hricak H: Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016; 278:563–5773.
3. Alamo L, Anaye A, Rey J, Denys A, Bongartz G, Terraz S, Artemisia S, Meuli R, Schmidt S: Detection of suspected placental invasion by MRI: Do the results depend on observer’ experience? Eur. J. Radiol. 2013; 82:e51–e57