4551
Video C3D features learned by deep network correlate with functional MRI signal variation associated with the video1Center for Advanced Neuroimaging, UC Riverside, Riverside, CA, United States, 2Dept. of Electrical & Computer Engineering, UC Riverside, Riverside, CA, United States, 3Dept. of Psychology, UC Riverside, Riverside, CA, United States, 4Dept. of Bioengineering, UC Riverside, Riverside, CA, United States
Synopsis
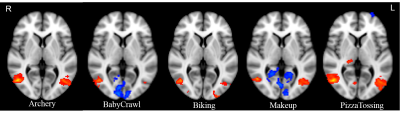
To gain further insights into the mechanisms of deep network learning from the perspective of brain imaging, we compared spatio-temporal features of video segments extracted via a 3-dimensional convolutional network (3D ConvNets) with video representations in human brain characterized by functional MRI signal variation when viewing video segments. We found correlations between C3D features and fMRI signal variation in brain regions selectively activated by video segments after the optimization of time lag due to the hemodynamic response function (HRF). Distinct activation patterns were also revealed by functional MRI for video segments classified as different classes of activity.
Introduction
Despite the tremendous success of deep learning networks in video computing in the past several years, many questions remain regarding the mechanism of their exceptional performance and how to achieve the optimal. C3D feature learned with 3-dimensional convolutional networks (3D ConvNets) is the state-of-the-art spatio-temporal feature for video as it works as a generic video descriptor solving large-scale video tasks.1 Here we explored the association between C3D features of video and functional MRI signal variation associated with video viewing to gain deeper insights into the mechanisms of 3D ConvNets from the perspective of brain imaging.Methods
Ten subjects (7 Male, 26.3±6.8 years old) were studied using Siemens Prisma 3T. Structural data were collected using a T1-MPRAGE sequence with the following parameters: TR/TE/TI=2400/2.72/1060ms, FA=8°, spatial resolution=0.8mm isotropic. BOLD functional MRI were acquired using a 2D GRE EPI sequence with the following parameters: TR/TE=750/37ms, FA=56°, slice thickness=2mm, in-plane resolution=2×2mm, multiband factor=8. During the functional scan, subjects were instructed to view a video consisting of 50 segments. These video segments were classified into 5 classes (10 segments for each), each corresponding to a certain activity: archery, baby crawling, biking, applying makeup and tossing pizza dough. The average video length (mean(SD)) for each class is 5.59(0.66) s, 5.69(1.73) s, 6.72(1.40) s, 7.61(2.36) s and 4.43(0.87) s, respectively. These video segments were concatenated in random order and interleaved with 10s transitions in between. The length of the entire video was 785s and was played at a frame rate of 25 fps.
Neuroimage analysis:
All data were processed using FSL. At the individual level, fMRI data were pre-processed with motion correction, slice timing correction, brain extraction, spatial smoothing and high pass filtering with a cut-off of 100s. The fMRI images were registered to the high resolution T1 using FLIRT with a boundary-based-registration (BBR) and then to the MNI template using FNIRT. General Linear Model (GLM) was setup to explore the activation associated with video segments from each class.
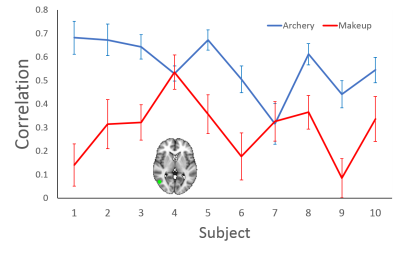
GLM with FLAME was used to obtain the group activation over 10 subjects for each activity class. A 6 mm spherical ROI centered at the peak group activation in the occipital cortex was delineated. The ROI was then mapped back to individual functional space and the average time series for that ROI was generated for each subject.
C3D feature extraction:
For each of the video segments, convolutional 3D (C3D) features of size 4096 were calculated over each 16 frames with a temporal stride of 4 frames. Principal component analysis (PCA) was then applied to the C3D feature and the 1st principal component (PC) was used for subsequent correlation analysis. We used only the 1st PC because it accounts for a majority amount of total variance (archery=88.8%(5.1%), baby crawl=86.7%(6.3%), biking=80.2%(3.0%), makeup=90.4%(5.4%), pizza tossing=85.0%(3.8%)).
Pearson correlation was then calculated between the 1st PC of C3D feature of each video segment and the average ROI time series optimized for the time lag due to the hemodynamic response function.