4511
Neuropharmacological fMRI of MDMA – A Novel, Multimodal Analytical Approach Informed by PET1Department of Neuroimaging, Institute of Psychiatry, Psychology & Neuroscience, King's College London, London, United Kingdom
Synopsis
This study evaluates the degree to which resting-state fMRI (rfMRI) response indexes the action at drug target sites by testing whether haemodynamic response to MDMA mirrors receptor 5-HT density profiles measured with PET. We weighted the rfMRI BOLD signal using a high-resolution in vivo atlas of the serotonin system. Results show that an altered haemodynamic response to MDMA is detectable only in the maps related to MDMA serotonin targets. This study provides new evidence that rfMRI haemodynamic response to MDMA reflects the known binding profile of the drug and set the basis for a biologically-informed rfMRI analysis in drug challenges.
Introduction
In the last decade, resting state fMRI (rfMRI) has been increasingly used in pharmacological applications to investigate drugs effects on brain function1. Pharmacological MRI studies strongly rely on the assumption that haemodynamic measures can be considered a proxy of altered neurotransmission function. However, the haemodynamic MRI signal has no intrinsic selectivity to any particular receptor sites. Indeed, the degree to which the functional MRI response indexes the action at drug target sites, and thus could be useful in parsing mechanistic underpinnings, is still an open question. The aim of this work is to address this question with a compound with a known mixed profile of serotonergic action. Specifically, we test whether haemodynamic response to 3,4-Methylenedioxymethamphetamine (MDMA) measured with rfMRI mirrors receptor 5-HT density profiles measured with PET.Methods
Twenty healthy subjects participated in this placebo-controlled crossover study. Images were acquired under two different conditions, i.e. placebo and 3,4-Methylenedioxymethamphetamine (MDMA), with a 3T scanner. rfMRI was acquired with a multi-echo EPI sequence (TR=2500ms, TEs=12, 28, 44ms, resolution = 3.75×3.75×4.2 mm3, slice thickness=3 mm, 27 axial slices; 192 volumes). High-resolution T1-weighted images were also acquired. rfMRI images were preprocessed with AFNI2 and de-noised with the AFNI tool meica.py3-5. Then, data were spatially smoothed with a with an 8-mm FWHM Gaussian kernel, WM and CSF signals were regressed out and a high-pass temporal filter, with a cut-off frequency of 0.005 Hz, was applied. Finally, images were normalized to MNI space using the Advanced Normalization Tools (ANTs)6. MDMA has its highest affinity for serotonin 1A, 2A receptors and the transporter7. We used a high-resolution in vivo atlas8 of four serotonin receptors (5-HT1A, 5-HT1B, 5-HT2A, and 5-HT4) and its transporter (5-HTT) as template in a two-step multivariate regression analysis9 to estimate the subject-specific spatial maps and time series of the BOLD response to placebo and MDMA. For each 5-HT target, the spatial maps of the two conditions were then compared using permutation tests with Randomise10.Results
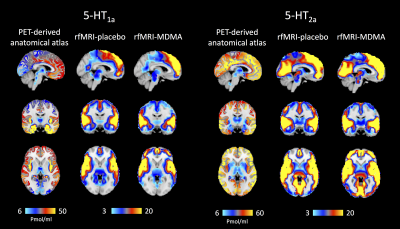
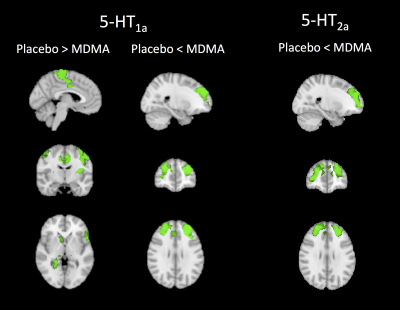
Figure 1 shows the PET density maps of the MDMA serotonin targets, i.e. the 5-HT1A and 5-HT2A receptors, and the corresponding resting state fMRI maps averaged across subjects for both conditions. By comparing the two conditions for every 5-HT target, significant differences were found only for the maps derived by two of the drug targets, namely the 5-HT1A and 5-HT2A receptors (pFWE < 0.05, figure 2). Specifically, subjects under MDMA showed a lower BOLD response (5-HT1A-related maps) in the precentral and postcentral gyri, supplementary motor cortex, insular cortex, anterior cingulate cortex, inferior frontal gyrus and subcortical structures (right thalamus, right caudate, right and left putamen, right accumbens and right hippocampus). A higher BOLD response to MDMA was also found in the frontal pole, middle frontal gyrus, paracingulate gyrus and anterior cingulate cortex (5-HT1A and 5-HT2A maps). Of note, a trend-level difference between the two conditions (placebo > MDMA, p = 0.0504) was found for the maps derived by the 5-HTT map.Discussion
The areas showing a significant different haemodynamic response to placebo/MDMA are those reported in published ASL and fMRI studies11,12. Moreover, significant differences between placebo and drug were found only in the maps determined by 5-HT1A and 5-HT2A density profiles. This result is in keeping with the strong affinity of MDMA for these two receptors7. The decreased BOLD response to MDMA in the 5-HTT-related maps, albeit at a trend level, is also in line with previous PET studies that reported a decreased global and regional brain 5-HT transporter binding in MDMA users13.Conclusion
This study provides new evidence that rfMRI changes following MDMA administration reflect the known binding profile of the drug. Ongoing work is validating this approach with other compounds (i.e. antipsychotics, psilocybin, ketamine) including the density profiles of other receptors (e.g. dopamine, glutamate) and other MRI modalities (i.e. ASL). This approach may provide an interesting new fingerprint in the characterisation of novel compounds and potentially greater insight to the commonly observed eclectic response to treatment.Acknowledgements
This abstract represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.References
1. Khalili-Mahani N, Rombouts SA, van Osch MJ, Duff EP, Carbonell F, Nickerson LD, et al. Biomarkers, designs, and interpretations of resting-state fMRI in translational pharmacological research: A review of state-of-the-Art, challenges, and opportunities for studying brain chemistry. Hum Brain Mapp. 2017;38(4):2276-325. doi: 10.1002/hbm.23516. PubMed PMID: 28145075.
2. Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29(3):162-73. PubMed PMID: 8812068.
3. Kundu P, Santin MD, Bandettini PA, Bullmore ET, Petiet A. Differentiating BOLD and non-BOLD signals in fMRI time series from anesthetized rats using multi-echo EPI at 11.7 T. NeuroImage. 2014;102:861-74.
4. Kundu P, Brenowitz ND, Voon V, Worbe Y, Vertes PE, Inati SJ, et al. Integrated strategy for improving functional connectivity mapping using multiecho fMRI. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(40):16187-92. doi: 10.1073/pnas.1301725110 [doi].
5. Dipasquale O, Sethi A, Lagana MM, Baglio F, Baselli G, Kundu P, et al. Comparing resting state fMRI de-noising approaches using multi- and single-echo acquisitions. Plos One. 2017;12(3):e0173289. doi: 10.1371/journal.pone.0173289. PubMed PMID: 28323821; PubMed Central PMCID: PMCPMC5360253.
6. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033-44. doi: 10.1016/j.neuroimage.2010.09.025. PubMed PMID: 20851191; PubMed Central PMCID: PMCPMC3065962.
7. Lyon RA, Glennon RA, Titeler M. 3,4-Methylenedioxymethamphetamine (MDMA): stereoselective interactions at brain 5-HT1 and 5-HT2 receptors. Psychopharmacology (Berl). 1986;88(4):525-6. PubMed PMID: 2871581.
8. Beliveau V, Ganz M, Feng L, Ozenne B, Hojgaard L, Fisher PM, et al. A high-resolution in vivo atlas of the human brain's serotonin system. J Neurosci. 2016. doi: 10.1523/JNEUROSCI.2830-16.2016. PubMed PMID: 27856786.
9. Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A. 2009;106(17):7209-14. doi: 10.1073/pnas.0811879106. PubMed PMID: 19357304; PubMed Central PMCID: PMCPMC2678478.
10. Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. Permutation inference for the general linear model. Neuroimage. 2014;92:381-97. doi: 10.1016/j.neuroimage.2014.01.060. PubMed PMID: 24530839; PubMed Central PMCID: PMCPMC4010955.
11. Carhart-Harris RL, Wall MB, Erritzoe D, Kaelen M, Ferguson B, De Meer I, et al. The effect of acutely administered MDMA on subjective and BOLD-fMRI responses to favourite and worst autobiographical memories. Int J Neuropsychopharmacol. 2014;17(4):527-40. doi: 10.1017/S1461145713001405. PubMed PMID: 24345398.
12. Walpola IC, Nest T, Roseman L, Erritzoe D, Feilding A, Nutt DJ, et al. Altered Insula Connectivity under MDMA. Neuropsychopharmacology. 2017;42(11):2152-62. doi: 10.1038/npp.2017.35. PubMed PMID: 28195139; PubMed Central PMCID: PMCPMC5603811.
13. McCann UD, Szabo Z, Scheffel U, Dannals RF, Ricaurte GA. Positron emission tomographic evidence of toxic effect of MDMA ("Ecstasy") on brain serotonin neurons in human beings. Lancet. 1998;352(9138):1433-7. PubMed PMID: 9807990.
Figures