4357
Automatic Segmentation of Lung Anatomy from Proton MRI based on a Deep Convolutional Neural Network1University of Virginia, Charlottesville, VA, United States, 2University of Missouri School of Medicine, Columbia, MO, United States
Synopsis
With rapid development of pulmonary MRI techniques, increasingly useful morphological and functional information can be obtained, such as pulmonary perfusion, ventilation and gas uptake through hyperpolarized-gas MRI. Identification of lung anatomy is usually the first step for quantitative analysis. In the work, we proposed and validated a new approach for automatic segmentation of lung anatomy from proton MRI based on 3D U-Net structure. The new method had a relatively consistent performance in all subjects (dice overlap 0.90-0.97). Its future application for anatomical based analysis of structural and/or functional pulmonary MRI data needs further validation in larger number of data.
Introduction
With rapid development of MRI techniques, increasingly useful morphological and functional information can be obtained from pulmonary MRI, such as pulmonary perfusion1, ventilation2 and gas uptake through hyperpolarized-gas MRI3,4. Identification of lung anatomy is usually the first step for quantitative analysis. Different automatic proton pulmonary segmentation approaches5,6 have been proposed in the past, and achieved moderate to relatively high accuracies. Most recently, deep neural networks have emerged to show great potential in image classification and segmentation for many applications. In this work, we propose and validate a new approach for automatic segmentation of lung anatomy from proton MRI based on the 3D U-Net structure7, a convolutional neural network that has already demonstrated good performance in biomedical image segmentation.Methods
The atlas dataset from ref. 6, which included 62 annotated proton MRI lung image sets from 48 subjects (24 females, 9 healthy, 19 with chronic obstructive pulmonary disease [COPD], 20 asthmatics), was used to train the U-Net for segmenting the whole lung and to separate the left and right lungs. This U-Net network took 3D volumes as input data, and all processing operations, such as convolution, pooling and de-convolution, were implemented in the corresponding 3D architecture. Our model is similar with the structure in ref. 7 but is optimized for this target data set and task. The network consisted of fourteen 3x3x3 convolutional layers interleaved by three 2x2x2 max-pooling in the down-sampling path and three 2x2x2 deconvolution layers in the up-sampling path. The features from the down-sampling path are fused with the corresponding up-sampling layers through concatenations and additional convolutions. In the end, the output labels are classified as background, left or right lung. After training, a segmentation mask volume was achieved for each test image through the network.
The performance of this method was evaluated in eight subjects (4 healthy subjects [H1-4], age: 59 ± 5 yrs; and 4 patients with COPD [C1-4], age: 61 ± 5yrs) against a “ground truth” dataset manually annotated by two experienced scientists familiar with lung MRI. In addition, we compared the performance of this new method to that proposed in ref. 6 on the same test dataset. For MRI data collection, each subject underwent 3-D gradient-echo based proton MRI acquisition with an acceleration factor of three. Generation of under-sampling patterns and image reconstruction followed descriptions in ref. 8. Other MR pulse sequence parameters were isotropic resolution= 3.9 mm, TR= 1.80 ms, TE= 0.78 ms, flip angle= 10o, bandwidth= 1090 Hz/Pixel.
Results and Discussion
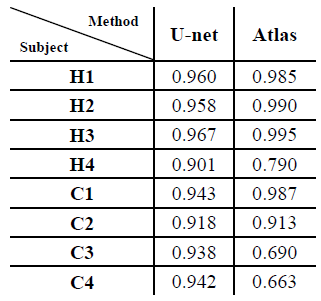
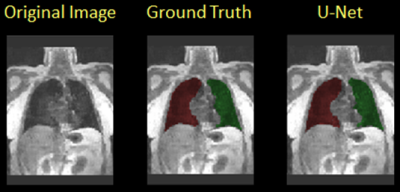
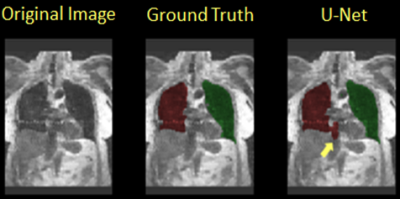
Table 1 shows the Dice overlap with expert manual annotation in the eight subjects participating in this study for the proposed method based on U-Net (‘U-Net’, left) and the previous method described in ref. 6 (‘Atlas’, ‘right’). The new method had an overall more consistent performance among all subjects (Dice overlap 0.90-0.97). In subject H1-3 and C1 the Atlas-based method had a slightly better performance (>0.98). A segmentation example for the U-Net method is shown in Figure 1. In subjects H4, C3 and C4, with more prominent imaging artifacts due to undersampling, the proposed method showed a more robust performance. Of note is that in some subjects, the current method could not distinguish some surrounding organs or tissues from lung areas if they have similar signal intensities and the boundaries separating them from lung areas were not very clear. One example of this is shown in Figure 2 (yellow arrow). A portion of the inferior vena cava, which was very close to the base of the left lung and had similar signal intensity on this MR image, was mistakenly treated as part of the left lung. Further refinement of the model, such as by adding appropriate shape constraints and expanding the training data set with more advanced data augmentation, are currently under way to improve the performance of this method. Also, additional functionality, such as lobar segmentation, will be implemented in the near future.Conclusion
In this study, a new method for whole lung segmentation based on 3D U-Net was proposed and tested in eight subjects who underwent proton lung MRI. Its future application for anatomically-based analysis of structural and/or functional pulmonary MRI data needs further validation in a larger data set.Acknowledgements
This work is supported by R21HL129112, R01HL109618, R01HL133889, R01HL132287, and R01HL132177 from the National Heart, Lung, and Blood Institute and research funding from Siemens Medical Solutions.References
[1].Ley S, et. Al. Insights Imaging. 2012 Feb; 3(1): 61–71. [2]. Woodhouse N, et. Al. J Magn Reson Imaging. 2005 Apr; 21(4): 365-9. [3]. Qing K, et. Al. J Magn Reson Imaging. 2014 Feb; 39(2): 346-359. [4]. Kaushik SS, et. Al. Magn Reson Med.. 2016 Apr; 75(4): 1434-1443. [5].Kohnmann P, et. Al. Int J Comput Assist Radiol Surg. 2015 Apr; 10(4): 403-17. [6].Tustison NJ, et. Al. Magn Reson Med. 2016 Jul; 76(1): 315-20. [7]. Ronneberger O, et. Al. arXiv:1505.04597. [8].Qing K, et. Al. Magn Reson Med. 2015 Oct; 74(4): 1110-15.Figures