4334
Diagnostic Assessment of Breast Cancer in Non-Contrast MRI Images Through an Artificial Intelligence Machine Learning Algorithm1Anatomy & Neurobiology, Boston University School of Medicine, Boston, MA, United States, 2Radiology, Boston Medical Center, Boston, MA, United States
Synopsis
Contrast-Enhanced Breast MRI is a common method for diagnosis of Breast Cancer. An Artificial Intelligence Machine Learning Algorithm was developed to analyze Non-Contrast Breast MRI scans and predict diagnoses. The algorithm was trained using MRI data that had pathological specimens to validate diagnoses, obtained from The Cancer Imaging Archive. The AI was found to be 95% accurate in classifying tissue as cancerous or benign. This algorithm could be used to assist diagnoses in clinical practice. Future work will assess the generalizability of the algorithm on data from other scan sites, and the potential for classifying specific subtypes of Breast Cancer.
Introduction
Breast MRI offers advantages over mammography by not exposing patients to ionizing radiation, but relies on administration of intravenous Contrast Agents1. Artificial intelligence (AI) machine learning algorithms for Computer Assisted Diagnosis are software packages that intake labelled images of different classes (i.e. cancerous images and benign images) and create statistical functions that explain the variance between classes2. AI uses learning theories and training algorithms which mimic techniques used in clinical training. AI offers a way to aid clinicians in diagnostic accuracy by applying quantitative algorithms on individual pixel values of images2,3,4,5. This study sought to develop an AI approach to accurately and efficiently differentiate cancerous from benign tissue.Methods
Non-contrast T2-weighted Turbo spin echo MRI images of 21 Patients with confirmed Breast Cancer (n = 21) and Patients without Breast Cancer (n = 9) were obtained from the BREAST-DIAGNOSIS6 collection of The Cancer Imaging Archive7, totaling 2,766 independent slices. All cancer diagnoses had corresponding pathological specimens for validation. An AI system using a Convolutional Neural Network (CNN) was built through Python scripting language to accurately identify specific imaging characteristics. The framework used was Google’s™ TensorFlow© 1.3.0, built to utilize the Graphical Processing Unit (GPU). A gradient descent algorithm was used to train the algorithm and minimize the loss function. The pipeline accepts both DICOM and NIfTI file types as inputs, although the BREAST-DIAGNOSIS data is in DICOM format. DICOM volumes were converted to single-slice JPEG images and labelled as cancerous or benign. Validation accuracy was calculated as the percentage of calculated classifications that matched pathological diagnosis, using a 10% holdout threshold. Images were randomly shuffled and parsed into either training (90%) or testing (10%) data groups. Images that were used to train the algorithm were not used to test the algorithm. Efficiency was determined in seconds per slice4. Our algorithm was trained and assessed using the unprocessed, native images that were acquired.Results
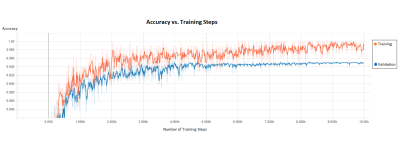
Using our neural network AI, tissue was classified as cancerous or benign with a validation accuracy of 95%. The algorithm was trained for 10,000 steps at a rate of 100ms per step, meaning that the algorithm was trained, tested, then re-trained with tuned parameters 10,000 times to increase accuracy (Figure 1). The AI classifies images at a rate of 2.28 seconds/slice, and automatically iterates through each slice of a volume.Discussion
Other CNN’s using TensorFlow have shown diagnostic accuracies up to 60.9% in classifying mammograms8. Our study has shown a diagnostic accuracy of up to 95% in pathologically-validated subset of Patients using Breast MRI. One of the challenges in any quantitative analysis of medical images is the variability in patient anatomy and image acquisition parameters3,5,9. The data used in this project were all collected using the same 1.5T Philips© Achieva™ MRI scanner and TSE SENSE™ pulse sequence. This simplified our analysis, since we did not need to correct for differences in image weighting. However, this also means the generalizability of the algorithm to other scan sites is untested. Our classifier was constructed to differentiate slices as cancerous or benign, but could potentially be trained to specifically identify types of cancer (e.g., invasive ductal carcinoma vs. invasive lobular carcinoma). This could expand the algorithm’s clinical usefulness, if successful. We used a high-powered workstation to train and assess the neural network; the Windows™ PC contained 64GB of RAM, a CPU overclocked to 4.1GHz, and an 8GB GeForce© GTX 1080™ GPU. A workstation equipped with lower-grade hardware would not process as quickly, but theoretically would produce the same results. It is common practice to pre-process images before assessment with a machine learning algorithm, using techniques such as segmentation, smoothing or normalization5,9,10,11. However, any pre-processing techniques change the image and could potentially introduce new features, or obfuscate existing features, in the data11. The decision not to pre-process the images may have increased the algorithm’s accuracy on this dataset.Conclusion
CNN offers a novel way to accurately identify Patients with Breast Tumors using non-contrast MRI images. This has important clinical applications for non-invasive Breast Cancer detection, diagnosis and assessment, as well as differentiation of tumor subtypes, facilitating optimized treatment decisions and optimizing treatment planning for the individual patient. AI algorithms can be trained to detect individual tumor characteristics, allowing clinicians to identify aspects of the tumor and its individual behavior prior to treatment and for disease monitoring5,11. Further work can assess the ability to specifically classify tumor types, the effect of pre-processing on classification accuracy, and the algorithm’s generalizability to other scan sites and pulse sequences.Acknowledgements
We would like to acknowledge The Cancer Imaging Archive, and specifically the team from the BREAST-DIAGNOSIS study, for curating this dataset and making it freely available to the public. Without their efforts, we would not have been able to complete this research.References
1. Mann, R. M., Kuhl, C. K., Kinkel, K., & Boetes, C. (2008). Breast MRI: guidelines from the European Society of Breast Imaging. European Radiology, 1307-1318.
2. Kleesiek, J., Biller, A., Urban, G., Kothe, U., Bendszus, M., & Hamprecht, F. A. (2014). ilastik for Multi-modal Brain Tumor Segmentation. Proceedings MICCAI BraTS (Brain Tumor Segmentation Challenge), 12-17.
3. Dalmis, M. U., Litjens, G., Holland, K., Setio, A., Mann, R., Karssemeijer, N., & Gubern-Nerida, A. (2017). Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Medical Physics, 533-546.
4. Zhou, X., Kano, T., Koyasu, H., Li , S., Zhou, X., Hara, T., . . . Fujita, H. (2017). Automated Assessment of breast tissue density in non-contrast 3D CT images without image segmentation on a deep CNN. Proceedings of SPIE.
5. Sun, W., Tseng, T.-L., Zhang, J., & Qian, W. (2016). Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Computerized Medical Imaging and Graphics, 4-9.
6. Bloch, B. Nicolas, Jain, Ashali, & Jaffe, C. Carl. (2015). Data from BREAST-DIAGNOSIS. The Cancer Imaging Archive.
7. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057.
8. Zhou, H., Zaninovich, Y., & Gregory, C. (2017, November 8). Mammogram Classification Using Convolutional Neural Networks. Retrieved from GitHub: http://ehnree.github.io/documents/papers/mammogram_conv_net.pdf
9. Nie, K., Chen, J.-H., Chan, S., Chau, M.-K. I., Yu, H. J., Bahri, S., . . . Su, M.-Y. (2008). Development of a quantitative method for analysis of breast density based on three dimensional breast MRI. Medical Physics, 5253-5262.
10. Lele, C., Wu, Y., DSouza, A. M., Abidin, A. Z., Xu, C., & Axel, W. (2017). MRI Tumor Segmentation with Densely Connected 3D CNN.
11. Kamnitsas, K., Ferrante, E., Parisot, S., Ledig, C., Nori, A. V., Criminisi, A., . . . Glocker, B. (2017). DeepMedic for Brain Tumor Segmentation. Brain lesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, 138-149.