4234
Imaging cardiac fat by chemical shift encoded MRI application in CINE format at 3 T1Philips Healthcare, Beijing, China
Synopsis
Ectopic fat deposition impacts cardiac health, and non-invase quantification is required. In this work, a chemical shift encoded MRI is designed for cardiac applications. Images of Proton density fat fraction (PDFF) water (W) and fat(F), and T2* were generated for each frame of cardiac motion, formed CINE movies for cardiac cycles. The novel sequence was flexible in clinical applications with short acquisition time and made sensitive findings in this research.
Purpose
Ectopic fat deposition has been associated with cardiac dysfunctions (1). Echocardiography, Epicardial adipose tissue (EAT) is a biologically active organ that may play a role in the association between obesity and coronary artery disease (CAD) (2), and is a predictor of coronary atherosclerosis. Pericardial adipose tissue (PAT) is useful in predicting factor of newincidences of paroxysmal atrial fibrillation (3). Cardiac tomography and some cardiac MR application have been developed for cardiac fat quantitative assessment. However, it remains a challenging task to robustly measure cardiac fat and further implement for clinical applications due to respiratory and cardiac motion. Chemical shift encoded MRI (CSE-MRI) for determining fat quantification has been validated and implemented for liver and other organ fat quantifications (4,5). Hereby we evaluate the feasibility in using CSE method for cardiac fat quantification, both in epicardial adipose tissue (EAT) and pericardial adipose tissue (PAT).
Considering periodicity of heart motion, this cardiac CSE MRI is implemented in CINE format and generate images at each cardiac phase.
Materials and Methods
Modified CSE-MRI sequence for cardiac CINE images was developed on a 3T MRI unit (Philips 3.0T Ingenia, Philips Healthcare, the Netherlands) with 6 gradient echoes as the number used for validated liver fat quantification application (mDIXON quant). Balanced sequence was designed with single slice to get bright blood effect. 4 chamber view protocol was designed with TR = 7.6ms, and TE1=1.36ms with echo spacing 1ms. The image acquisition used 144×203 matrix with voxel size of 2.08mm×1.48mm. Slice thickness 8mm, TFE factor 6 with 6 echoes, compressed sense acceleration factor at 3, and total scan time 11.6s. ECG cardiac trigger was applied to acquire 21 images. short axis view protocol was designed 1.17×1.17mm resolution with 152×187 matrix. TR 6.4ms TE1 1.19ms with echo spacing 0.8ms. and result in 24 phases.
Magnitude and phase images at 6 gradient echo are generated at each of the cardiac phases, forming a 6×21 matrix. Proton density fat fraction (PDFF) along with water (W) and fat(F) images, and T2* images were calculated based on the 6 complex numbered images. Then PDFF, W, F and T2* each forms frames cardiac CINE.
This Institutional Review Board (IRB) was approved for this study. Volunteers and a patient suspected with coronary artery disease underwent cardiac CSE-MRI.
Results
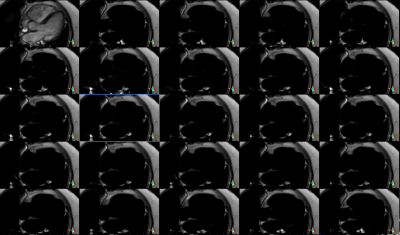
CINE images from volumteers were shown in figure 1, with good contrast in water.
Also fat only images could be displayed in CINE and form movies. Such as shown in figure 2.
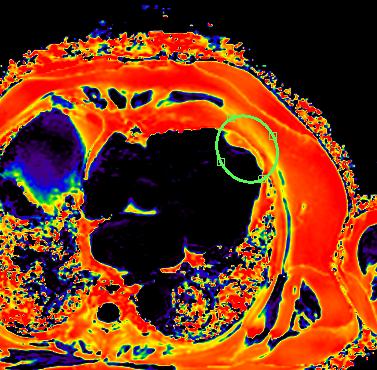
An spot on proton density fat fraction CINE images was found for the patient, cardiac SWI scan also found this abnormity. CT images, which were taken in a month, were retrieved and reconstructed to look into the EAT at the apical region to compare with CSE-MRI results in figure 3.
Conclusion and Discussion
Fat quantification in epicardial adipose tissue (EAT) or pericardial adipose tissue (PAT) are important for heart viability assessment and a good biomarker for coronary artery diseases. The advances cardiac applications such as chemical shift encoded MRI (CSE-MRI) is feasible. And the set of images at each cardiac phase sort by echo number and cardiac phase can generate proton density fat fraction (PDFF) along with water (W) and fat(F) images, and T2* images.
This study conclude the feasibility in applying CSE-MRI cardiac scans. With the fast imaging methods such as compressed SENSE, the challenges from respiratory and cardiac motion are substantially reduced. And more resolution and contrast will be achieved with optimized CSE-MR for cardiac applications.
Acknowledgements
No acknowledgement found.References
- Daniel Davidovich, Amalia Gastaldelli, Rosa Sicari; Imaging cardiac fat, European Heart Journal - Cardiovascular Imaging, Volume 14, Issue 7, 1 July 2013, Pages 625–630,
- Talman AH, Psaltis PJ, Cameron JD, Meredith IT, Seneviratne SK, Wong DT. Epicardial adipose tissue: far more than a fat depot. Cardiovasc Diagn Ther 2014;4(6):416-429. doi: 10.3978/j.issn.2223-3652.2014.11.05 Okura, Kiyotaka & Hirazawa, Motoaki. (2016).
- PERICARDIAL ADIPOSE TISSUE AS A MORE USEFUL PREDICTING FACTOR OF NEW INCIDENCES OF PAROXYSMAL ATRIAL FIBRILLATION THAN THE SEVERITY OF CORONARY ARTERY DISEASE. Journal of the American College of Cardiology. 67. 1771. 10.1016/S0735-1097(16)31772-7.
- Kukuk, GM. et al., Comparison between modified Dixon MRI techniques, MR spectroscopic relaxometry, and different histologic quantification methods in the assessment of hepatic steatosis. European Radiology, 2015 Apr 23.
- Kijowski, R., et al., Improved fat suppression using multipeak reconstruction for IDEAL chemical shift fat-water separation: Application with fast spin echo imaging. Journal of Magnetic Resonance Imaging, 2009. 29(2): p. 436-442.