4190
Deep Learning Enables 90% Reduction in Gadolinium Dosage for Contrast Enhanced MRI1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Synopsis
There are increasing concerns over gadolinium-based-contrast-agents-administration(GBCA). A deep-learning (DL) method was developed to reduce the gadolinium dose in Contrast-Enhanced-MRI (CE-MRI). The proposed method includes an acquisition step(pre-contrast, 10% low-dose and full-dose CE-MRI with T1-weighted-IR-FSPGR), a pre-processing step and a deep learning model trained to predict full-dose CE-MRI from pre-contrast and low-dose images. Evaluated on a clinical neuro CE-MRI dataset (10 patients for training and another 20 patients for evaluation), both quantitative metrics and radiologists’ ratings showed the proposed method achieved improved synthesis, with better motion-artifact-suppression and NO significant differences in contrast-enhancement quality, compared with ground-truth full-dose CE-MRI. Thus, using the proposed Deep Learning method, GBCA can be reduced, by at-least-10-fold, while preserving image quality and diagnostic information.
Introduction
MRI is a powerful imaging technique providing unique tissue contrasts to distinguish different tissue types and pathologies. Gadolinium based contrast agents (GBCA) are frequently used to enhance MRI and boost the visibility of lesions[1]. However there are increasing concerns for GBMA administration because of a number of side effects including allergy reactions[2], nephrogenic systemic fibrosis (NSF)[3] and, the most importantly, gadolinium deposition[4]. In this work, we develop and validate a novel deep learning (DL)[5] technology to significantly reduce GBCA dose levels while maintaining necessary diagnostic quality for clinical interpretations.Method
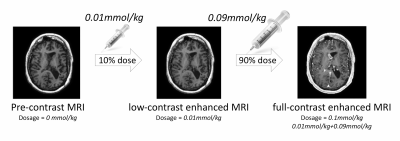
Imaging Protocol: High-resolution 3D T1-weighted inversion-recovery prepped fast-spoiled-gradient-echo imaging (IR-FSPGR, T1 BRAVO, TR/TE/TI 9.5/3.8/400ms, matrix 512x512, FOV 240mm, thickness 1mm) was collected using 3T MRI scanners (GE Healthcare) on 30 patients ($$$48.5 \pm 19.2$$$ years, 17 males) receiving clinically indicated CE-MRI with clinical protocol. The protocol, as shown in Figure 1, includes following 3 sequences: high-resolution pre-contrast, post-contrast images with 10% low-dose and 100% full-dose of gadobenate dimeglumine (0.01 and 0.1 mmol/kg, respectively).
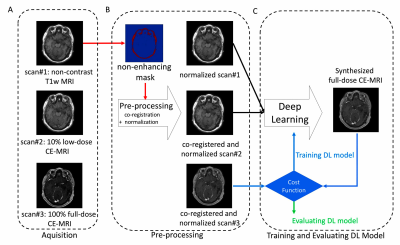
Pre-processing: The workflow of the method is shown in Figure 2. To remove systematic differences in non-enhancing regions and mitigate the variability of transmit and receive gains, co-registration and signal normalization was conducted based on average voxel value within masks.
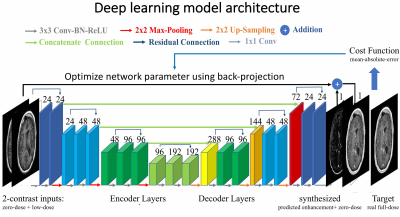
Training Deep Learning model: The detailed model architecture (Figure 3) is an encoder-decoder convolutional neural network (CNN) with bypass connections and residual connections as U-Net[5,6]. The deep network implicitly learns the guided denoising of noisy contrast uptake in low-dose CE-MRI to synthesize full-dose CE-MRI from pre-contrast and low-dose CE-MRI. We trained the model using the first 10 cases and evaluated the trained model on the other 20 patients. In the training we used Mean-Absolute-Error cost function and the ADAM[7] method.
Assessment: The acquired low-dose, real full-dose, and the synthesized full-dose post-contrast images were compared quantitatively using peak-signal-to-noise-ratios (PSNR), structural-similarity-indexes (SSIM) and qualitative scores by 2 neuroradiologists.
Results
Based on the quantitative similarity metrics, the proposed DL method yielded significant (p<0.001) improvements over the $$$10\%$$$ low-dose images, with over 5dB PSNR gains, $$$11.0\%$$$ SSIM increases and significant improvements in qualitative ratings.
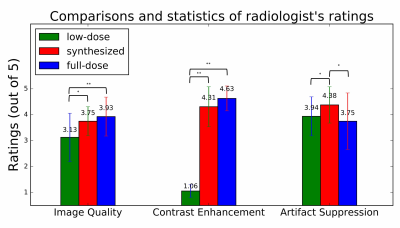
From radiologists’ ratings (Figure 4), there were no significant differences (both p>0.05) in image quality or contrast enhancement between the synthesized and acquired full-dose images. In addition, significantly better (p<0.05) artifact suppression was noted.
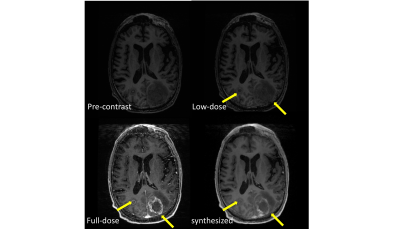
Figure 5 shows a visual comparison between pre-dose MRI, 10% low-dose CE-MRI, reference full-dose CE-MRI and the synthesized CE-MRI using the proposed method. The synthesized image demonstrates high image quality, preserved contrast enhancements and less motion artifacts.
Discussion
The proposed deep learning based method enables a 90% reduction of Gadolinium dose, which significantly reduces risks for patients. It can also be used to enhance image quality for cases where low contrast dose has already been administrated for reasons (e.g. pediatric imaging).
The proposed technique can be further improved by using more controlled experiment designs to mitigate the effects of different imaging settings and timing. In addition, further performance improvement can be achieved with 3D models and different cost functions[8]. It is reasonable to explore further reduction of Gadolinium dosage above the 90% reduction, by incorporating improved DL algorithms, using larger training datasets with more variable pathologies as well as other complimentary information from multi-contrast MRI and clinical history.
Conclusion
With the proposed DL based method, the proposed method can at least reduce 90% of the gadolinium dose in CE-MRI, while maintaining good image quality and preserving enhanced contrast for diagnosis, which is of great benefit to patients.Acknowledgements
No acknowledgement found.References
- Brasch RC, Weinmann HJ, Wesbey GE. Contrast-enhanced NMR imaging: animal studies using gadolinium-DTPA complex. American Journal of Roentgenology. 1984 Mar 1;142(3):625-30.
- Prince MR, Zhang H, Zou Z, Staron RB, Brill PW. Incidence of immediate gadolinium contrast media reactions. American Journal of Roentgenology. 2011 Feb;196(2):W138-43.
- Marckmann P, Skov L, Rossen K, et al. Nephrogenic systemic fibrosis: suspected causative role of gadodiamide used for contrast-enhanced magnetic resonance imaging. Journal of the American Society of Nephrology. 2006 Sep 1;17(9):2359-62.
- Gulani V, Calamante F, Shellock FG, Kanal E, Reeder SB. Gadolinium deposition in the brain: summary of evidence and recommendations. The Lancet Neurology. 2017 Jul 31;16(7):564-70
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. InInternational Conference on Medical Image Computing and Computer-Assisted Intervention 2015 Oct 5 (pp. 234-241). Springer, Cham.
- Chen H, Zhang Y, Kalra MK, Lin F, Liao P, Zhou J, Wang G. Low-Dose CT with a Residual Encoder-Decoder Convolutional Neural Network (RED-CNN). arXiv preprint arXiv:1702.00288. 2017 Feb 1.
- Ngiam J, Coates A, Lahiri A, Prochnow B, Le QV, Ng AY. On optimization methods for deep learning.In Proceedings of the 28th international conference on machine learning (ICML-11) 2011 (pp. 265-272).
- Mardani M, Gong E, Cheng JY, Vasanawala S, Zaharchuk G, Alley M, Thakur N, Han S, Dally W, Pauly JM, Xing L. Deep Generative Adversarial Networks for Compressed Sensing Automates MRI. arXiv preprint arXiv:1706.00051. 2017 May 31.
Figures