4167
A Three-dimensional Template Matching Technique in Target Tracking Technique of MRgHIFU for liver1Graduate School of System Informatics, Kobe University, Kobe, Japan, 2Information Science and Technology Center, Kobe, Japan, 3Graduate School of Engineering, Tokai University, Hiratsuka, Japan, 4Center for Frontier Medical Engineering, Chiba University, Chiba, Japan
Synopsis
MRgHIFU treatment for liver requires a tracking technique to “lock on” to the focal spot at the target tissue region during respiratory-induced motion. We proposed three-dimensional focus tracking technique using template matching method. In this method, by creating a deformity / mutation model reconstructed from multi-slice volume data and extracting multiple blood vessel branch points, the focal spot can be estimated from the relative positional relationship. In this study, we optimized template size of template matching for extracting branching points in preoperative. Experimental results demonstrated to improve accuracy of blood vessel extraction.
Purpose
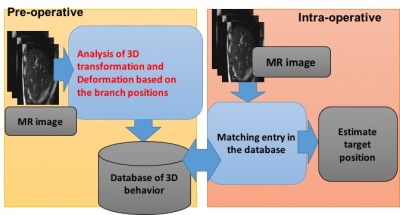
MRgHIFU treatment of a liver requires a tracking technique to “lock on” to the focal spot at the target tissue region during respiratory-induced motion for targeting and temperature imaging. A method using a blood vessel branching structure considering three-dimensional deformation has been proposed [1][2]. In this method, a database relating to the movement of the liver is prepared before the operation(Figure 1). In the database, information on the position of the target using the relative positional relationship between the target and surrounding blood vessel branches is included. In order to extract the movement of a vessel branch from the acquired pre-operative images, a template matching method using vessel branching as a region of interest is used. Here, accurately extracting the position of blood vessel branch points is important in improving accuracy of focus estimation. In this study, we investigated ideal conditions of template matching for extracting blood vessel branch points in preoperative.Materials and Methods
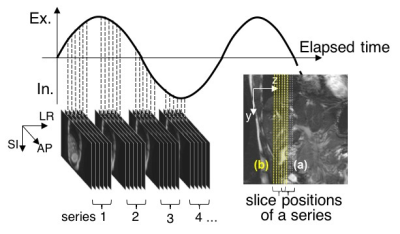
MR data acquisition: Two 42-image series at intervals of 15 mm, which included 6 interleaved sagittal images from a healthy volunteer ’s liver, were acquired by a 3.0T MRI with steady-state free precession (SSFP) under natural slow breathing and a single coronal slice was acquired under breath holding (Figure 2). Imaging conditions were follows: TR/TE, 4.85ms/1.98ms; FA, 90 degrees; slice thickness, 5mm; FOV, 350x350mm2; acquisition matrix, 256x256; parallel factor, 2.0.
Reconstruction of a 3D image series: First, the diaphragm positions of each image were extracted with the 3D template matching method [2]. Template patterns were set in the area that includes the diaphragm in each first image series. The sum of the absolute difference (SAD) was used for the evaluated value of dissimilarity between the template and the input pattern. Next, each image series was classified into four categories (inhalation, maximal inhalation, exhalation, and maximal exhalation), depending on relative displacement, with the diaphragm position of the coronal slice as a reference image. The images of each slice position were sorted in respiratory order. The sorted images were interpolated in the direction of the respiration cycle and assembled into image sets in such a way that the diaphragm positions fit to the diaphragm line of the coronal image. Each assembled image set was rearranged toward the isotropic voxel images via a linear interpolation method.
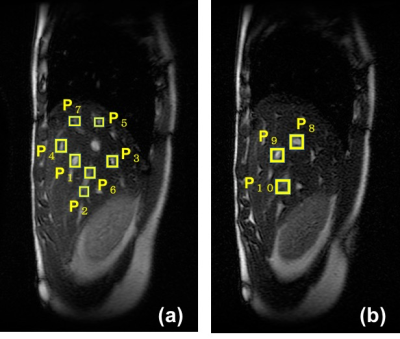
Vessel branch extraction: The vessel branch positions of each image were extracted with the 3D template matching method. First, The sufficiently small rectangle templates which contained each vessel branch point were set as shown in Figure 3. The sum of the absolute difference (SAD) and zero-mean normalized cross correlation(ZNCC) were used for evaluated value of similarity between the template and the input pattern. To save the time, the image area for examining similarity was limited to the range that could be moved by respiration. The evaluated values of similarity were repeatedly searched by dilating the templates one pixel at a time. And the errors between extracted position and the right position set by manual operation were calculated.
Results and Discussion
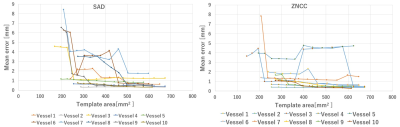
Figures 4 are the relationship between the average error of each extracted vessel branch point and the area size of the template. The correct solution position of the vessel branch points was set to the average value of a plurality of extraction results with errors eliminated by visual observation. Figure 4 shows that by setting the area of the template to 500 mm2 or more, error was less than 2 mm in all vessel extraction. Therefore, as the size of the template was set large, the error did not decrease but increased due to the influence of parts that did not change due to breathing. Also, since the time and the size of the template size were a trade-off, it was desirable that the size be small.Conclusion
In this study, we optimized size of the template and found that highly accurate tracking was possible by setting the template area to 500 mm2 or more for the current data. In preoperative processing, extracting branch points with high precision leads to highly accurate tracking of the focal position, and it is the same as being able to perform intra-operative tracking with higher accuracy. High precision tracking is indispensable for clinical application.Acknowledgements
This research is supported by grant number 17K01357 from the JSPS Grant-in-Aid for Scientific Research (C).References
1) Kumamoto E, Matsumoto T, Hayashi M, and Kuroda K, Four-dimensional Reconstruction of Abdominal MR Images for Target Tracking Based on Respiration-induced Liver Trans- formation and Deformation for MR-guided Focused Ultrasound Surgery, JJME, Vol53, No 3, pp.168–178, 2015.
2) Kumamoto E, Matsumoto T, Kokuryo D, Kuroda K, Three-dimensional target tracking method for MRgHIFU using image matching technique with liver deformation volumes, Proc of 44th JSMRM, pp.245, 2017.
Figures