4103
Sliding Motion Corrected Low-rank Plus Sparse Reconstruction for Free-breathing Liver DCE-MRI1College of Information Science and Electronic Engineering, Zhejiang University, Hangzhou, China, 2Department of Radiology, Weill Cornell Medical College, New York, NY, United States, 3Department of Biomedical Engineering, Cornell University, Ithaca, NY, United States
Synopsis
Liver dynamic contrast enhanced MRI (DCE-MRI) requires high spatiotemporal resolution to clearly visualize enhancement patterns. Image reconstruction quality is often compromised due to unavoidable respiratory motion during the long-time acquisition. This abstract presents a novel sliding motion corrected low-rank plus sparse (SMC-LS) reconstruction algorithm for free-breathing liver DCE-MRI. The sliding motion of the liver along the superior-inferior direction is estimated directly from the under-sampled k-space data. Low-rank and sparse regularization are enforced on the sliding motion corrected image reconstructions. Results demonstrated that SMC-LS can substantially reduce motion blurring and preserve more details in free breathing liver DCE-MRI.
PURPOSE
Dynamic contrast enhanced MRI (DCE-MRI) with high spatiotemporal resolution and large volume coverage enables the accurate diagnosis of liver lesions in their early stages. Previously proposed reconstruction methods, such as blind compressive sensing1, TRACER2, PROUD3, and low-rank plus sparse (L+S)4, can achieve satisfactory image quality for motion-free k-space data which are usually acquired using multiple breath-holds protocol, but fails for free breathing data with larger respiratory motion. Recently, XD-GRASP5 demonstrated promising performance at 11-12 seconds frame rate for free breathing liver DCE-MRI. In this study, we investigated a new method which incorporates respiratory Sliding Motion Correction into conventional L+S reconstruction (SMC-LS) such that motion blurring artifacts could be minimized while retaining a high temporal frame rate.METHODS
In SMC-LS, the liver 4D DCE-MRI reconstruction
problem is formulated as
$$ \min _{L_i,S_i} \ \sum_{i=1}^{n_z}(\lambda_{L}||L_i||_* + \lambda_{S}||TS_i||_1)+\frac{1}{2}|| E( \tau \sum_{i=1}^{n_z}(R_i(L_i+S_i)))-D||_2^2 $$
where the first part exploits low rank and sparsity of the reconstructed image, and the second part enforces data consistency. $$$n_z$$$ is the number of axial slices,$$$L_i$$$ is the low-rank component and $$$S_i$$$ is the sparse component of axial slice i in the 4D image after inter-frame sliding motion registration,$$$T$$$ is the total variation operator which enforces sparsity, $$$\lambda_L$$$ and $$$\lambda_S$$$ are regularization parameters.$$$R_i$$$ is an operator that puts the motion corrected slice image,$$$L_i+S_i$$$ ,back into a 4D image. $$$E$$$ denotes the k-space data acquisition operator, which performs a multiplication by multi-channel coil sensitivities followed by an under-sampled Fourier transform, and $$$D$$$ denotes the acquired raw data.$$$\tau$$$ is a motion operator which models the frame-by-frame image transformation driven by respiration. In free breathing, the major component of liver motion is its sliding along the superior-inferior (SI) direction, whereas the abdominal wall moves a little bit inwards and outwards, and the back bone stays stationary. In SMC-LS, $$$\tau$$$ is simplified to one dimensional sliding motion of inner organs. Based on the acquired k-space center data of axial slices, inter-frame sliding motion in the SI direction is estimated by one dimensional data registration and coil clustering6. A combination of singular value thresholding used for matrix completion7 and iterative soft thresholding used for sparse reconstruction8 is used to solve the above minimization problem.
For the validation of SMC-LS, we reused one case of in vivo liver DCE-MRI data acquired in PROUD3 with single breath-hold protocol. New in vivo data were acquired in another volunteer with IRB approval using the same parameters: 1.5T scanner, 8-channel cardiac coil, multiphase spiral LAVA with 48 golden angle variable density spiral leaves per fully sampled volume, voxel size 1.48×1.48×5 mm3, and matrix size 256×256×36. A total of 4 volumes (192 spiral leaves) were continuously acquired using free breathing protocol but without contrast injection. Free breathing liver DCE-MRI data were artificially made by firstly transforming the breath-hold liver DCE MR images with the sliding motion signal derived from the free breathing case and then acquiring the k-space data with the E operator. XD-GRASP5 and conventional L+S4 reconstruction were also tested for comparison.
RESULTS
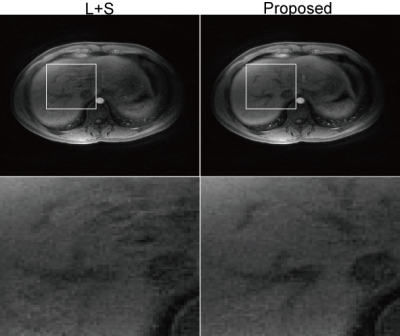
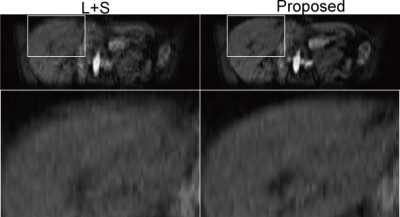
For L+S and SMC-LS, one frame was reconstructed using every two continuous spiral leaves, resulted in totally 96 frames at about 2 frames/s and. For XD-GRASP, two configurations were tested: 6 DCE states by 6 motion states, and 4 DCE states by 4 motion states. Fig. 1 shows a comparison of reconstructed images of L+S and SMC-LS in axial sections with zoomed-in ROIs, and Fig. 2 shows an image comparison in coronal sections with zoomed-in ROIs. Compared to the convention L+S, motion blurring artefacts were significantly reduced, and image details were better preserved by the proposed method. Fig. 3 shows a comparison of reconstructed images of XD-GRASP and SMC-LS. Compared to the proposed method, XD-GRASP reconstruction with 36 states has apparent under-sampling artefacts, while XD-GRASP reconstruction with 16 states were temporally smoothed due to lower temporal resolution.
DISCUSSION
Low rank and sparsity are prone to be corrupted by inter-frame misalignment, resulted in compromised image quality. The preliminary data presented here show the feasibility of enhancing conventional L+S reconstruction with respiration motion correction to overcome motion blurring in free breathing liver DCE-MRI. The proposed SMC-LS reconstruction simplifies the complex non-rigid respiratory motion to one dimensional sliding motion, which is estimated directly from the raw k-space data. By incorporating the inter-frame sliding motion correction, low rank and sparsity can be well recovered.CONCLUSION
Sliding motion corrected L+S reconstruction is demonstrated to be effective for preserving both spatial structures and high temporal resolution in free breathing liver DCE-MRI.Acknowledgements
We acknowledge support from NIH grants RO1 CA181566, RO1 EB013443 and RO1 NS090464.References
[1] Lingala SG, et al., TMI, 32(6): 1132-1145, 2013. [2] Xu B, et al., MRM, 69(2): 370-381, 2013. [3] Cooper MA, et al., MRM, 74(6): 1587-1597, 2016. [4] Otazo R, et al., MRM, 73(3): 1125-1136, 2015. [5] Feng L, et al., MRM, 75(2): 775-788, 2016. [6] Zhang T, et al., MRM, 76(1): 197-205, 2016. [7] Cai JF, et al., SIOPT, 20(4): 1956-1982, 2010. [8] Daubechies I, et al., COMMUN PUR APPL MATH, 57(11): 1413-1457, 2004.Figures