4096
Accelerating cardiac dynamic imaging with video prediction using deep predictive coding networks1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States
Synopsis
Deep predictive coding networks based on stacked recurrent convolutional neural network have shown great success in video prediction since they can learn to recognize and analyze the motion patterns of each element from previous frames. In this study we adopted this model to predict future frames in cardiac cine images and used a k-space substitution method to improve the prediction accuracy. It showed promises in accelerating cardiac dynamic imaging.
Introduction
With recent developments in deep convolutional neural networks, future frames in a video sequence can be predicted well using predictive networks 1,2 since they can learn to recognize and analyze the motion patterns of each element from previous frames. The deep predictive coding networks proposed in 2 (PredNet) use a stacked recurrent convolutional neural network (RCNN) to predict future frames and showed state-of-the-art performance compared with other network structures. In this study, we aim to adopt PredNet to predict future frames in cardiac cine images and explore the combination with k-space sampling to further improve the prediction accuracy.Methods
The network structure is similar to that described in Figure 1 of 2 with 4 stacked RCNN layers. The convolutional LSTM block in each layer is used to store information from previous frames. The numbers of features in each layer are 1, 48, 96 and 192 and the image size is reduced by 2x2 for each layer on top. The training and validation data set is from the Kaggle Second Annual Data Science Bowl 3 containing 2D short-axis images covering LV from 700 subjects. Each subject contains 8-12 slices with 30 cardiac phases for each slice. 500 subjects were used for training and 200 were used for validation. The model input is a dynamic image series containing 8 consecutive frames randomly extracted from the entire cardiac cycle and the output is the predicted next frames given all previous ones (e.g. predict frame 6 from frame 1-5). Root mean square error between the ground truth frame and the predicted frame is used as the loss function during training. During validation, all 30 cardiac phases are fed into the model to generate predicted next frames. To evaluate the model, the predicted next frames are compared with the corresponding ground truth frames and the previous frames, since a straightforward prediction method is just to copy the previous frame. To study the effect of using under-sampled k-space data, a simulation study was performed by transforming the ground truth frame and the predicted frame to k-space and substituting the k-space lines from the predicted frame with the corresponding lines from the ground truth frame according to the sampling pattern, which is 4x uniform undersampling in this experiment. The updated prediction is then made by an inverse FFT of the resulting k-space data. As a matter of fact, this operation, when applied to the simple prediction method by copying a previous frame, is the view sharing reconstruction, since the un-acquired k-space data is directly copied from a previous frame.Results
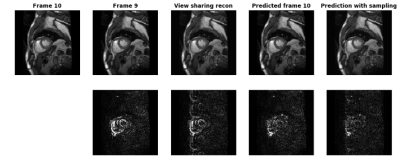
Figure 1 shows the results focusing on frame 10 at systole. The top row shows ground truth frame 10, frame 9, view sharing reconstruction, predicted frame 10 and the updated prediction using the k-space substitution method. The bottom row shows the differences with the ground truth frame. During systole, when cardiac motion is the fastest, there are significant changes around the myocardium between frame 9 and frame 10. The view sharing recon updates the cardiac shape but suffers from aliasing. With just model prediction, the mismatch in LV myocardium is greatly reduced, because the model learned the movement patterns of myocardium and predicted its deformation from the previous frames. With k-space substitution, the differences with ground truth are further minimized.Discussions and Conclusion
Through training from a large number of cardiac cine images, the PredNet model is able to learn the movement patterns on a pixel-by-pixel level to predict the next frame in the image series from previous frames. This prediction can be improved with k-space sampling by simply replacing the acquired k-space data. However, there are several limitations in this study. Due to the lack of original k-space data, the sampling step assumes the phase is zero in all images. In practice, if only the magnitude image series are available, one solution is to separate the real and imaginary parts of the image series and run the trained model on each part and combine the results before k-space substitution. Furthermore, as the training and validation data has minimal breathing artifacts, it is unclear whether the model can track global heart motions during breathing. However, given the success of PredNet in predicting the movements of real-life objects, as the breathing movement pattern is much simpler than myocardium deformation, it is likely to learn the breathing motion with only a handful of training data. In conclusion, PredNet is a promising tool for fast and reliable accelerated dynamic imaging.Acknowledgements
No acknowledgement found.References
1. Mathiu M, Couprie C, Lecun Y. Deep multi-scale video prediction beyond mean square error. arXiv:1511.05440 [cs.LG]. 2015.
2. Lotter W, Kreiman G, Cox D. Deep predictive coding networks for video prediction and unsupervised learning. arXiv:1605.08104 [cs.LG]. 2016.
3. https://www.kaggle.com/c/second-annual-data-science-bowl
Figures