4091
A dynamic MR-signal model to capture 3D motion-fields at ultra-high frame-rate1Centre for image sciences, University Medical Centre Utrecht, Utrecht, Netherlands, 2Utrecht University, Utrecht, Netherlands
Synopsis
We present a framework that has the potential to capture non-rigid 3D motion at 50 Hz, hereby drastically accelerating state-of-the-art techniques. Our model directly and explicitly relates the motion-field to the k-space data and is independent of the spatial resolution, allowing for extremely high under-sampling. We illustrate proof-of-principle validations of our method through a simulation test and whole-brain 3D in-vivo measured data. Results show that the 3D motion-fields can be reconstructed from extremely under-sampled k-space data consisting of as little as 64 points, enabling 3D motion estimation at unprecedented frame-rates.
Introduction
Real-time estimation of three-dimensional human motion/deformation is crucial for applications such as MRI-guided radiotherapy and cardiac imaging. State-of-the-art dynamic MRI methods (e.g. navigators1, cardiac gating2 and low-rank compressed sensing3 aim at overcoming the inherently slow MR data encoding rate but are still far from achieving real-time, high-resolution, non-gated 3D acquisitions.
Here, we present a framework that has the potential to capture non-rigid 3D motion at 50 Hz, hereby drastically accelerating state-of-the-art techniques. Our model directly and explicitly relates the motion-field to the $$$k$$$-space data and is independent of the spatial resolution, allowing for extremely high under-sampling.
Preliminary numerical and in-vivo experimental tests show that reconstruction of an affine 3D motion-field is possible with as little as 64 $$$k$$$-space points.
Theory
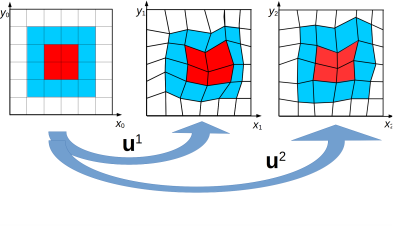
Our dynamic model directly relates the MR-signal of an object at time $$$t_{j+1}$$$ deformed by an unknown, possibly non-linear, motion-field $$$\boldsymbol{u}(\boldsymbol{r})$$$, to the reference image at time $$$t_j$$$ before deformation. The vector-field $$$\boldsymbol{u}$$$ thus induces a coordinate transformation between the two time points (Fig. 1):$$\boldsymbol{r}|_{t=t_j}\mapsto\boldsymbol{u}(\boldsymbol{r})|_{t=t_{j+1}}$$We assume that the magnetic spins are in the steady-state regime, thus\[m^{j+1}(\boldsymbol{r})=m^{j}(\boldsymbol{u}(\boldsymbol{r})).\] Here, the superscript indicates time and $$$m$$$ the (normalized) transverse magnetization. Additionally, we assume a spin-conservation relationship analogous to the mass-conservation:$$\rho^{j+1}(\boldsymbol{r})\text{d}\boldsymbol{r}=\rho^j(\boldsymbol{u}(\boldsymbol{r}))\text{d}\boldsymbol{u}(\boldsymbol{r})=\rho^j(\boldsymbol{u}(\boldsymbol{r}))\lvert J(\boldsymbol{u})\rvert\text{d}\boldsymbol{r}.$$Here, $$$\rho(\boldsymbol{r})$$$ denotes the spin density and $$$\lvert J(\boldsymbol{u})\rvert$$$ the determinant of the Jacobian of $$$\boldsymbol{u}$$$. Combining these assumptions and the change of variable formula for the multi-dimensional integral we arrive at the following formula for the signal at time $$$t_{j+1}$$$:$$s^{j+1}\left(\boldsymbol{k}\right)=\int_{\mathbb{R}^3}\rho^{j+1}(\boldsymbol{r})m^{j+1}(\boldsymbol{r})e^{-i 2\pi\boldsymbol{k}\cdot\boldsymbol{r}}\text{d}\boldsymbol{r}=\int_{\mathbb{R}^3}\rho^j(\boldsymbol{r})m^j(\boldsymbol{r})e^{-i 2\pi\boldsymbol{k}\cdot\boldsymbol{u}^{-1}(\boldsymbol{r})}\text{d}\boldsymbol{r}.$$In this work, we refer to $$$q^j(\boldsymbol{r}):=\rho^j(\boldsymbol{r})m^j(\boldsymbol{r})$$$ as the reference image and $$$s^{j+1}(\boldsymbol{k})$$$ as the snapshot data. Note that $$$\boldsymbol{u}(\boldsymbol{r})$$$ could represent non-linear/non-rigid transforms.Methods
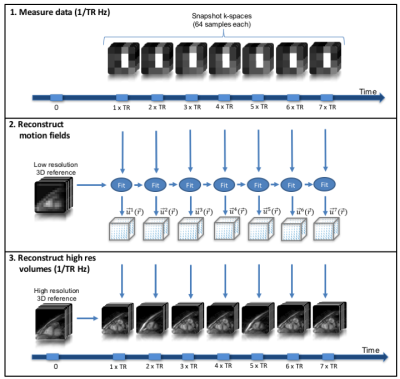
Given an MR-signal at time $$$t_{j+1}$$$ and a reference image at time $$$t_j$$$ we can reconstruct the deformation-field $$$\boldsymbol{u}(\boldsymbol{r})$$$ by numerically solving a non-linear inversion problem. See Fig. 2. We illustrate proof-of-principle validations of our method through a simulation test and whole-brain 3D in-vivo measured data.
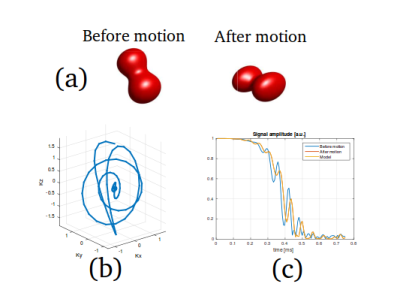
For the numerical test, we considered an affine transformation $$$\boldsymbol{u}(\boldsymbol{r})$$$, i.e. $$\boldsymbol{u}(\boldsymbol{r})=A\boldsymbol{r}+\boldsymbol{\tau},$$where $$$A\in\mathbb{R}^{3\times3}$$$ and $$$\boldsymbol{\tau}\in\mathbb{R}^3$$$. We employed a bSSFP sequence with a very short (sub-millisecond) 3D spiral readout trajectory (see Fig. 3). From the signal after motion (snapshot data) we reconstructed the affine transformation and compared the hereby deformed object with the ground-truth.
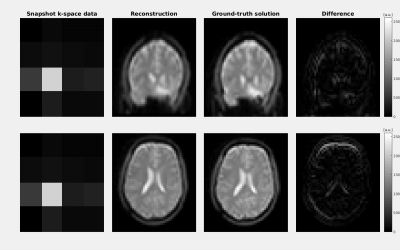
For the whole-brain 3D in-vivo test, we represented the motion-field by a non-rigid affine transformation. We acquired one reference image, and a total of seven snapshot $$$k$$$-spaces. The volunteer moved in between the scans. Data was acquired using a heavily under-sampled 3D steady-state SPGR sequence with Cartesian readout on a 1.5T MR system (Ingenia, Philips), TR/TE=8ms/3ms. Retrospectively down-sampling eventually resulted in a pre-motion reference image of size $$$34\times34\times34$$$ and $$$k$$$-space snapshots of size $$$4\times4\times4$$$ (i.e. 64 points acquired in $$$4\times4\times$$$TR=128ms). To obtain ground-truth images, we performed a 3D SPGR with a single-shot EPI readout (TR/TE=35ms/9ms) immediately after each snapshot scan.
Results
For the simulations, Fig. 3c shows that our model accurately predicts the true signal after motion. Fig. 4 shows the transformed object by applying the reconstructed motion-fields; a very high accuracy in the reconstruction is achieved.
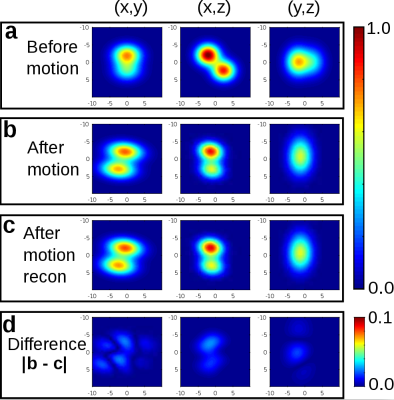
Results of the in-vivo experiment are reported in Fig. 5 and show good agreement between our reconstruction and the ground-truth. We used a plain channel-wise sum of raw MR-signal data for our snapshots, lacking any form of channel sensitivity corrections. This possibly explains the difference in our reconstruction.
Discussion
We introduced a framework to reconstruct non-rigid 3D transformations from minimal MR-signal data. This study presented simulated and experimental proof-of-principle results, showing the potential of the method. In the future, we will employ fast single-shot spiral acquisitions and expect to achieve a high-resolution 3D acquisition protocol with 50 frames per second (TR=20ms).
While the head motion is described well by affine transformations, other organs (abdomen and/or heart) will require more local and non-rigid parameterizations for $$$\boldsymbol{u}$$$; splines and other compressed representations derived from existing 4D human models4 will be considered. Note that our proof-of-principle affine transformation reconstructions are obtained with only 64 points of Cartesian $$$k$$$-space. More sophisticated transformations may require different trajectories and/or more $$$k$$$-space points but should still fit in the targeted 50 Hz rate.
The highly under-sampled data leads to a reduced minimization problem, which could be solved in less than a second with our initial Matlab implementation. Real-time reconstruction should be obtained by making use of CUDA/GPU implementations.
Conclusion
We derived a new MR-signal model which directly and explicitly relates the measured data to the motion/deformation-field. Simulations and experimental proof-of-principles show that these fields can be reconstructed from extremely under-sampled $$$k$$$-space data, enabling 3D motion estimation at unprecedented frame-rates.Acknowledgements
This research is funded by the Netherlands Organisation for Scientific Research, domain Applied and Engineering Sciences, Grant number: 15115.References
- Welch, E. B., Manduca, A., Grimm, R. C., Ward, H. A., and Jack Jr, C. R. Spherical navigator echoes for full 3D rigid body motion measurement in MRI. Magnetic Resonance in Medicine 47 , 1 (2002), 32–41.
- Uribe, S., Muthurangu, V., Boubertakh, R., Schaeffter, T., Razavi, R., Hill, D. L., and Hansen, M. S. Whole-heart cine MRI using real-time respiratory self-gating. Magnetic Resonance in Medicine 57 , 3 (2007), 606–613.
- Otazo, R., Candès, E., and Sodickson, D. K. Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components. Magnetic Resonance in Medicine 73 , 3 (2015), 1125–1136.
- Segars, W., Bond, J., Frush, J., Hon, S., Eckersley, C., Williams, C. H., Feng, J., Tward, D. J., Ratnanather, J., Miller, M., et al. Population of anatomically variable 4D xCAT adult phantoms for imaging research and optimization. Medical physics 40 , 4 (2013).
Figures