4088
A Comparison of Prospective versus Retrospective Motion Correction for Reliable T1-Weighted Neuroimaging1University of Wisconsin - Madison, Madison, WI, United States
Synopsis
Prospective motion corrected MPRAGE T1-weighted images were compared against retrospectively motion corrected MPnRAGE T1-weighted images. 14 young children were scanned without sedation and without the head being stabilized with pads. Dice-overlap of automatic tissue segementation masks (FAST and FLIRT) were used as a measure of reliability. MPnRAGE with motion correction showed exceptional regional label consistency (>=80% Dice overlaps for all 16 regions and >90% in 12 of the regions). Conversely, MPRAGE-PROMO demonstrated lower performance than even MPnRAGE without motion correction.
Introduction
Head motion in pediatric neuroimaging studies can lead to serious image artifacts, such as ghost-artifacts when Cartesian k-space sampling is used. Strategies to minimizing motion artifacts may reduce the need for sedation, which would be desirable for pediatric studies. Prospective motion correction (PROMO1) estimates head motion in near-real-time to adjust the imaging gradients for future TRs. PROMO has been applied to MPRAGE sequences to reduce motion artifacts for T1-weighted (T1w) imaging. An alternative T1w imaging method, MPnRAGE2 was used for retrospective motion correction using self-navigation properties of radial k-space sampling3,4.
In this study, the reliability of prospective motion correction of MPRAGE-PROMO is compared against MPnRAGE with and without self-navigated retrospective motion correction. An evaluation was performed using repeated T1w scans in a cohort of young children. The consistency of repeated automated image segmentation was used to assess the reliability of the image quality for repeated scans.
Methods
Fourteen children (109+/- 30 months, min=76 months ,max=166 months) were imaged at 3T using a 32-channel coil w/o sedation. Each subject was scanned three times in a single session with both MPRAGE-PROMO and MPnRAGE (six scans total). The first scan with each method was collected with foam padding to restrict head motion. The padding was then removed so that head motion was less impeded. Two additional MPnRAGE and MPRAGE-PROMO scans were performed in alternating order. The order of all scans was also counter-balanced across subjects
scan parameters (summary): Both methods acquired 3D whole head scans with 1.0 mm isotropic resolution. Scan times were matched at 7.0 minutes (MPRAGE-PROMO had 6.0 min scan + 1 min rescan). MPnRAGE produces multiple inversion time images (TIs), but only one with similar contrast to MPRAGE was chosen for analysis. MPnRAGE was reconstructed both with and without motion correction. MPnRAGE and MPRAGE images were coregistered within subject and processed with “fsl_anat” from the FMRIB Software Library v5.0. This includeded bias-field correction [FAST5], brain-extraction [FNIRT-based], tissue-type segmentation [FAST], and subcortical structure segmentation [FIRST6]. Dice-overlap-coefficients (DOCs) for each possible pair of similar scans were calculated for the white matter (WM), gray matter (GM) and cerebrospinal-fluid (CSF) whole brain masks from FAST and the 15 subcortical masks from FIRST.
The displacement of the center-of-mass between adjacent navigator images was calculated and used to assess the degree of motion for each scan.
Results
The mean of the absolute value of the displacements between the center-of-mass of adjacent navigator images for MPRAGE-PROMO and MPnRAGE scans was 0.4 for both methods, suggesting comparable motions across methods.
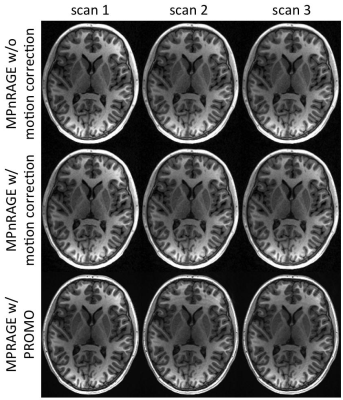
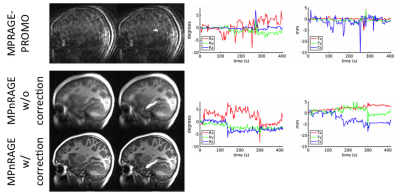
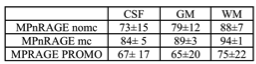
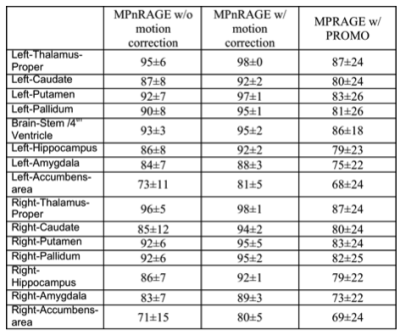
When motion was minimal, both methods performed similarly (Fig. 1), even though MPRAGE-PROMO showed mild ringing/motion artifacts. As the motion increased, MPnRAGE (with and without motion correction) resulted in higher Dice-overlaps due to less motion artifacts (Fig. 2). The Dice-overlap-coefficients are summarized in Figure 3-4.
Discussion
Overall, the automated image segmentation and anatomical labeling software tools showed more consistent performance with MPnRAGE than MPRAGE-PROMO. In particular, MPnRAGE with motion correction showed exceptional regional label consistency (>=80% Dice overlaps for all 16 regions and >90% in 12 of the regions). Conversely, MPRAGE-PROMO demonstrated lower performance than even MPnRAGE without motion correction. As both methods correct for motion at about the same rate (2.5s for MPRAGE-PROMO and 1.9s for MPnRAGE), one potential mechanism to explain the difference in performance is the type of residual motion artifacts from motion in-between navigator updates, which neither method corrects for. It is possible that the segmention algorithms are more sensitive to ghosting (MPRAGE-PROMO) than local blurring (MPnRAGE) artifacts.
It should be noted that this study was performed in a young pediatric sample without stabilizing the head with padding, which is particularly challenging so the degrees of motion may be larger than encountered in most neuroimaging studies. However, we expect MPnRAGE with motion correction to demonstrate high consistency in nearly all cases, regardless of age or degree of motion.
Conclusions
In a challenging young pediatric sample, this study showed that retrospective motion correction with radial-based MPnRAGE produced much more consistent automated brain segmentations than Cartesian-based MPRAGE-PROMO. This is strong supporting evidence that MPnRAGE with motion correction will be an effective T1-weighted imaging method for studies in young children without sedation or other populations where motion is expected to be problematic. Further, the high consistency of image segmentation and anatomical labeling is very promising for future quantitative neuroimaging studies.Acknowledgements
Funding Sources:
GE Healthcare
UW-Madison Department of Radiology
NIH grants: MH100031, MH097464, AG051216, HD090256
References
1. White, N., Roddey, C., Shankaranarayanan, A., Han, E., Rettmann, D., Santos, J., Kuperman, J. and Dale, A. (2010), PROMO: Real-time prospective motion correction in MRI using image-based tracking. Magn. Reson. Med., 63: 91–105. doi:10.1002/mrm.22176
2. Kecskemeti, S., Samsonov, A., Hurley, S. A., Dean, D. C., Field, A. and Alexander, A. L. (2016), MPnRAGE: A technique to simultaneously acquire hundreds of differently contrasted MPRAGE images with applications to quantitative T1 mapping. Magn. Reson. Med., 75: 1040–1053. doi:10.1002/mrm.25674
3. Kecskemeti, S., Alexander A., (2014) Motion Corrected Radial MPnRAGE. ISMRM 2014, #4343
4. Alexander A., Kecskemeti, S., (2015) Retrospective Motion Correction of MPnRAGE Studies in Children. ISMRM 2015 #0833
5. Zhang, Y. and Brady, M. and Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imag, 20(1):45-57, 2001.
6. Patenaude, B., Smith, S.M., Kennedy, D., and Jenkinson M. A Bayesian Model of Shape and Appearance for Subcortical Brain NeuroImage, 56(3):907-922, 2011.
Figures