4025
Exploring Glutamate and Glutamine Metabolism Across Breast Cancer Subtypes using High-resolution Proton MRS Combined with Molecular Approaches1Russell H. Morgan Department of Radiology, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
We have studied glutamine metabolism in a panel of breast cancer cell types to examine the utilization of glutamine as an energy source in different subtypes of breast cancer. We utilized high-resolution 1H MRS detection of glutamine and glutamate from cell extracts to determine up regulation of glutamine metabolism as a whole followed by quantitative RT-PCR to begin to establish the critical pathways involved in altered glutamine metabolism within malignant cell types. Altered glutamine metabolite signatures may be used to identify malignancy within cells and key enzymes in glutamine metabolism may prove to be potential therapeutic targets.
Purpose
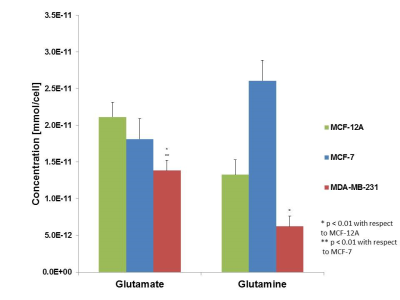
Glutamine, a non-essential amino acid, has been demonstrated as a major energy source across various cancers.1 Many types of tumors have demonstrated increased uptake of glutamine which serves as a carbon and nitrogen source as well as for shuttling essential amino acids into tumor tissue. This so-called “glutamine addiction” has been demonstrated across multiple cancer types including breast cancer, however, the mechanism of increased glutamine uptake has not been rigorously explored across the various subtypes, including nonmetastatic and metastatic, as compared to non-malignant cell lines.2 Cellular glutamine and glutamate levels can be monitored utilizing 1H MRS, and are detected at 2.34 and 2.45 ppm respectively, as previously demonstrated.3 In this study, we have investigated the glutamine profiles across multiple breast cancer cell lines via 1H MRS and explored the mechanism of glutamine uptake via computational methods and quantitative RT-PCR.Methods
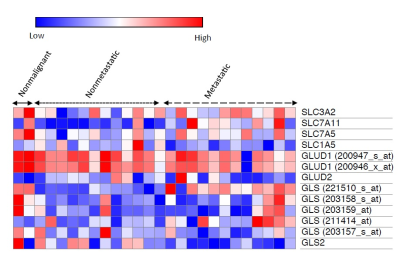
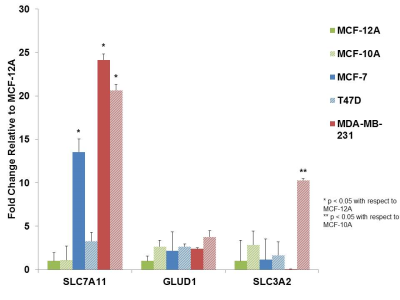
Results
Discussion
Conclusions
Glutamine/glutamate metabolism may play an important role in the aggressive nature of breast cancer, through the utilization of glutamine as a carbon and nitrogen source. Furthermore, glutamate/glutamine metabolism may be stimulated by the overexpression of glutamate transporters in malignant cells. Altered glutamate/glutamine metabolite signatures may be used to identify malignancy within cells and key enzymes in glutamate/glutamine metabolism may prove to be potential therapeutic targets. Studies in our lab are ongoing to further unravel the altered glutamine/glutamate metabolism in relation to breast cancer aggressiveness.
Acknowledgements
We thank all members of the Division of Cancer Imaging Research in The Russell H. Morgan Department of Radiology and Radiological Science for their help and support.References
1. Souba, WW. Glutamine and Cancer. Annals of Surgery, 1993. 218(6): 715-728.
2. Wise, DR., Thompson, CB. Glutamine Addiction: A New Therapeutic Target in Cancer. Trends Biochem. Sci., 2010. 35(8):247-433.
3. Chan KW, Jiang L, Cheng M, Wijnen JP, Liu G, Huang P, van Zijl PC, McMahon MT, Glunde K. CEST-MRI detects metabolite levels altered by breast cancer cell aggressiveness and chemotherapy response. NMR Biomed. 2016 Jun;29(6):806-16.
Figures