4013
NMR-BASED METABOLOMIC ANALYSIS AS A DIAGNOSTIC TOOL FOR TUBERCULOSIS IN CLINICAL URINE SAMPLES1CNIC, Madrid, Spain, 2CIBERES, Madrid, Spain, 3CNIO, Madrid, Spain, 4Servicio de Microbiologia. Hospital Universitari Germans Trias i Pujol. Institut d'Investigació Germans Trias i Pujol. Universitat Autònoma de Barcelona, Barcelona, Spain, 5CIBERES, Barcelona, Spain, 6Universidad Complutense de Madrid, Madrid, Spain
Synopsis
The ability of diagnose the tuberculosis infection is an essential factor in the spreading control of tuberculosis. However, microscopic examination presents a low sensitivity and culture techinques require incubation times up to two months. This study aimed at developing a NMR-based metabolomic approach for the differential diagnosis of tuberculosis in urine samples. We examined samples from patients diagnosed of tuberculosis (n=19), other respiratory infection (n=25) and healthy controls (n=29). Unsupervised PCA provide a nearly perfect discrimination between the three groups. We identified 31 chemical shifts regions to develop predictive models for the diagnostic of tuberculosis, obteining an accuracy of 100%.
Introduction
The ability of diagnose the tuberculosis infection in its primary stages is an essential factor in the spreading control of tuberculosis. The reference microbiological methods for tuberculosis diagnosis are microscopic examination, culture and isolation of Mycobacterium tuberculosis. However, microscopic examination presents a low sensitivity (approximately 50%) and culture techinques in solid media are slow, they require incubation times up to two months. Although PCR techiques for genetic amplification provide rapid results, they require especialised personnel and a well equipped laboratory. In this context, metabolomic analysis has emerged in the last years as a potentially useful tool for the discrimination between different etiologies of respiratory infections. It was observed that patterns obtained with urine metabolites were different depending on the etiology of the respiratory infection of the patient -S. aureus, Coxiella burnetti, Haemophilus influenzae, Mycoplasma pneumoniae and even M. tuberculosis [1]. Metabolic pattern of urine and plasma of paediatric patients diagnosed with pneumonia were also evaluated allowing the sucesfully clasification between pneumonic and control individuals [2]. This study aimed at developing a Nuclear Magnetic Resonance (NMR)-based metabolomic approach for the differential diagnosis of tuberculosis in urine samples.Material and Methods
Urine samples from patients diagnosed of tuberculosis (n=19), other respiratory infection (n=25) and healthy controls (n=29) were examined using a Bruker Avance spectrometer operating at 16.4 T. Before NMR acquisition, urine samples were pH adjusted using a 0.2M Phosphate Buffer (pH=7.4) containing 0.3 mM TSP as internal reference. 1D proton NMR spectra were recorded using a NOESY pulse sequence. Standard solvent-suppressed spectra were grouped into 32,000 data points, averaged over 256 acquisitions. The free induction decay (FID) signals were multiplied by an exponential weight function corresponding to line broadening of 0.3 Hz. Spectra were referenced to the TSP singlet at 0 ppm chemical shift. NMR spectra were data-reduced to equal length integral segments (δ=0.04 ppm) and they were normalized to total sum of the spectral regions. Principal Component Analysis (PCA) was applied to identify metabolic differences between groups. Classificatory models of partial least squares discriminant analysis (PLS-DA) was developed for the diagnosis of Tuberculosis. For training purposes, the classification functions derived from the probability of belonging to each group were computed with a number of random testing subjects. These classification functions were used afterwards to classify the rest of subjects as an internal validation. This process was repeated 100 times with random permutations of the data to reduce type I errors. The percentages of correct classification were calculated as a measure of the model performance.Results
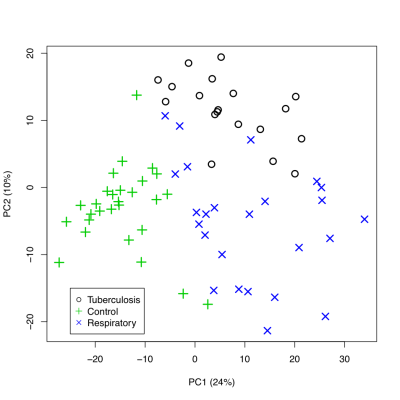
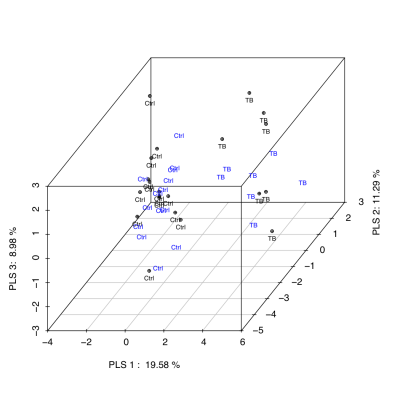
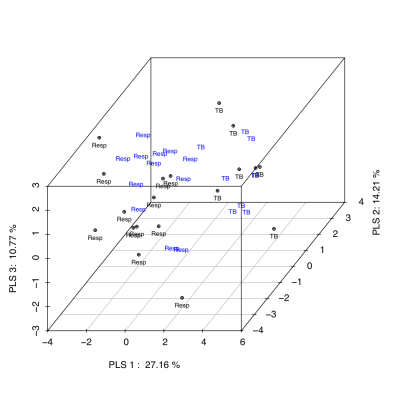
Unsupervised classification studies with PCA were carried out to analyze the differences between groups. The urine spectra provide a nearly perfect discrimination between the three groups along the first two principal components (Fig 1). PCA loading plots were used to highlighted the most significant variables. The potential biomarkers were selected by Hotteling’s T2 test. We identified 31 chemical shifts regions significantly diferent between groups. Using the selected chemical shifts, we developed predictive models for the diagnostic of tuberculosis. PLS-DA was applied to investigate the significant differences between tuberculosis patients and both respiratory infections and control groups. PLS-DA models were built with half set of samples, and were validated with other half set. Three latent variables (classification functions) were selected to build the PLS-DA models based on model robustness parameters, e.g. R2, Mean Squared Error of Prediction (MSEP) in Cross-Validation and Variance Explained. Both PLS-DA classification models, tuberculosis vs control (Fig 2) and tuberculosis vs respiratory infection (Fig 3), provided a diagnosis accuracy of about 100% by test samples.Discussion
In the present study, we show that the metabolomic profile obtained by NMR is sensitive to identify tuberculosis patients from healthy subjects and patients with other respiratory infections, thus being a potential biomarker for the specific diagnosis and prognosis of tuberculosisAcknowledgements
J.L.I.G is a CNIC IPP COFUND Fellow and has received funding from the People Programme (Marie Curie Actions) of the FP7/2007-2013 under REA grant agreement nº 600396. The CNIC is supported by MEIC-AEI and the Pro CNIC Foundation, and is a Severo Ochoa Center of Excellence (MEIC award SEV-2015-0505).References
[1]. Slupsky CM, Rankin KN, Fu H, Chang D, Rowe BH, Charles PG, et al. Pneumococcal pneumonia: potential for diagnosis through a urinary metabolic profile. J Proteome Res. 2009;8:5550-8.
[2]. Laiakis EC, Morris GA, Fornace AJ, Howie SR. Metabolomic analysis in severe childhood pneumonia in the Gambia, West Africa: findings from a pilot study. PLoS One. 2010;5.
Figures