3971
Spectral simulations of glutathione at 7T: Comparison of two different spin system parameter sets1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Biomedical Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
Glutathione (GSH) is a redox compound, providing protection against reactive oxygen species. Abnormal variation in GSH concentration has been associated with several neurological diseases. Several studies quantify low-concentration metabolites, such as GSH, using simulated basis sets derived from spin system parameters available in the literature. Through simulations and phantom experiments, we assess the accuracy of two different sets of spin system parameters of GSH-cysteine at 7T. The disagreement between the phantom and simulation GSH-cysteine spectra suggests a need for further refinement of the spin system parameters.
Purpose
Glutathione (GSH) is an important redox compound in the human brain, providing antioxidant protection against reactive oxygen species1. Abnormal GSH concentrations have been reported in several neurological diseases1. It is beneficial to quantify low-concentration metabolites, such as GSH, using magnetic resonance spectroscopy (MRS) at ultra-high-field (B0 ≥ 7T) because of higher signal-to-noise ratio and spectral dispersion2. Simulated basis sets are widely used for linear-combination modeling of proton (1H) MR spectra based on published tables of spin parameters, e.g. Govindaraju et al.3 and Tkac4. Because of the chemical shift differences between spins (in Hz) scale with B0 while the size of scalar couplings is independent of field strength, the degree of strong coupling in a spin system is field-strength-dependent. Here, we compare simulations performed with different GSH-cysteine spin system parameters3,4 to phantom experiments performed at 7T, assessing the degree of agreement and the need for further refinement of the spin system parameters.Methods
Phantom experiments were conducted on a Philips Achieva 7T scanner using a head coil with dual-channel transmit and 32 receive channels. Single-voxel MRS acquisitions were performed at a range of TEs using semi-localization by adiabatic selective refocusing (sLASER)5, comprised of a broadband frequency-modulated excitation pulse and two pairs of adiabatic pulses6 for refocusing. The bandwidth and duration of the excitation pulse (fremrex) were 4.73 kHz and 8.77 ms, respectively. The sweep width and duration of the adiabatic pulses (Rosenfeld/OIT) were 5 kHz and 5.23 ms, respectively.
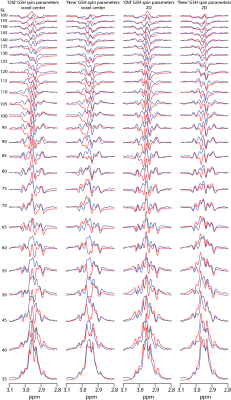
Simulation: 2-D density matrix simulations of the GSH-cysteine spin system were performed using FID-A7 at 7T for nominal voxel dimensions of 3 x 3 cm2. sLASER was simulated, assuming ideal excitation and shaped refocusing pulses (bandwidths as above), at TEs ranging from 35 to 160 ms in 5-ms increments using: 1. chemical shifts and coupling constants derived from3, simulating at the voxel center (non-localized); 2. chemical shifts and coupling constants derived from4 at the voxel center; 3. chemical shifts and coupling constants derived from3 on a 19 x 19 two-dimensional spatial array, as in reference8; and 4. chemical shifts and coupling constants derived from4 on the same spatial array. Spatially resolved simulations were performed in the dimensions defined by the refocusing pulses spanning 3.2 x 3.2 cm2. The simulations were performed with the following parameters at each position within the array: B1 = 15 μT; 8192 data points; 5 kHz spectral width; 2 Hz simulated linewidth; additional line broadening using a 3-Hz exponential filter. Transverse relaxation was simulated for a T2 of 70 ms.
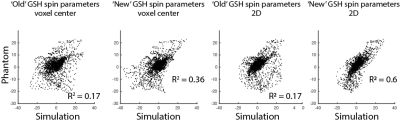
Phantom: A phantom was prepared at 25°C with pH of 7.2 in a phosphate-buffered solution containing 1.5 g/L NaN3 and 20 mM GSH and scanned on the same day. The same TE series was acquired in the phantom using the sLASER sequence with the following parameters: TR = 3000 ms; TEs = 35-160 ms in 5-ms increments; peak B1 = 15 μT; 5 kHz spectral width; 3 x 3 x 3 cm3 voxel; 16 averages per TE; and VAPOR water suppression. The phantom and simulation spectra, over the range 2.8 to 3.1 ppm, were plotted against each other and the coefficients of determination (R2) were calculated.
Results
GSH-cysteine simulations are overlaid with phantom spectra in Figure 1. Simulations using spin parameters from3 do not agree with the phantom spectra at several TEs. Simulations using spin parameters from4 show small improvements at the voxel center and improve further when simulated over the entire voxel, especially at TEs above 55 ms. The correlation plots are shown in Figure 2. The R2 is 0.17 for both the voxel center and the entire voxel using the parameters from3. The R2 is 0.36 for the voxel center and improves to 0.6 for the entire voxel using the parameters from4.Discussion
Simulations of GSH-cysteine disagree with the phantom experiments using the widely available spin parameters from3, but moderately agree using the new spin parameters4. It is likely that the remaining discrepancies arise from a combination of experimental and simulation imperfections. The mismatch between simulations and phantom is 7T-specific because simulations of GSH-cysteine at 3T are acceptable9. At 7T, the GSH-cysteine multiplet is highly impacted by strong coupling, with coupling becoming weaker at higher B0 and spins becoming increasingly equivalent at lower B0.Conclusion
Overall, the disagreement between simulation and phantom experiment suggests that further refinement of the published sets of spin system parameters, on which most in vivo spectral fitting is based, may be needed.Acknowledgements
This work was supported by NIH grants R01EB023963 and P41EB015909.References
1. Rae CD, Williams SR. Glutathione in the human brain: Review of its roles and measurement by magnetic resonance spectroscopy. Analytical biochemistry. 2017;529:127-43.
2. Deelchand DK, Marjańska M, Hodges JS et al. Sensitivity and specificity of human brain glutathione concentrations measured using short‐TE 1H MRS at 7 T. NMR Biomed. 2016;29(5):600-6.
3. Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129-53.
4. Tkac I, editor Refinement of simulated basis set for LCModel analysisIn Proceedings of the 16th Annual Meeting of ISMRM, Toronto, Canada, 2008.
5. Scheenen TW, Klomp DW, Wijnen JP, et al. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008 Jan;59(1):1-6.
6. Rosenfeld D, Zur Y. A new adiabatic inversion pulse. Magn Reson Med. 1996;36(1):124-36.
7. Simpson R, Devenyi GA, Jezzard P, et al. Advanced processing and simulation of MRS data using the FID appliance (FID‐A)—An open source, MATLAB‐based toolkit. Magn Reson Med. 2017;77(1):23-33.
8. Near J, Evans CJ, Puts NA, et al. J-difference editing of gamma-aminobutyric acid (GABA): simulated and experimental multiplet patterns. Magn Reson Med. 2013 Nov;70(5):1183-91.
9. Saleh MG, Oeltzschner G, Chan KL, et al. Simultaneous edited MRS of GABA and glutathione. Neuroimage. 2016;15:576-82.
Figures