3966
Non-water Suppressed GABA Edited Magnetic Resonance Spectroscopic Imaging using Density Weighted Concentric Rings k-space Trajectory1School of Health Sciences, Purdue University, West Lafayette, IN, United States, 2Wellcome Centre for Integrative Neuroimaging, FMRIB Division, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 3Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom, 4National Institute of Mental Health, National Institutes of Health, Bethesda, MD, United States, 5Department of Radiological Sciences, University of California, Los Angeles, CA, United States, 6Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, United States
Synopsis
In this study, we have developed and demonstrated a non-water suppressed GABA editing Magnetic Resonance Spectroscopic Imaging technique using density-weighted concentric rings k-space trajectory that performs robustly within a clinically feasible acquisition time at 3T. The method has been validated in a series of phantom experiments and its feasibility assessed in a healthy volunteer with a high in-plane resolution of 7.5 × 7.5 mm2. Experiments qualitatively demonstrate the advantage of the proposed method in terms of its improved resolution and reduced contamination of spectra from neighboring voxels.
Introduction
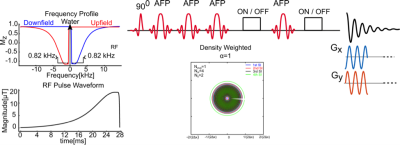
Water-suppressed MRS acquisition techniques have been the standard approach used in research and clinical MRI scanners to date. The acquisition of a non-water-suppressed spectrum is used for artifact correction, reconstruction of phased-array coil data, and metabolite quantification. A non-water suppressed metabolite-cycling (1) MRSI acquisition technique has recently been proposed for simultaneous detection of the metabolite and water signals. Potential improvements of voxel-wise frequency, phase, and eddy current correction have been demonstrated (1). In this study, we propose using a MEGA semi-LASER sequence with non-water suppressed metabolite-cycling as 2D MRSI data acquisition method at 3T for high-resolution GABA detection throughout a whole axial slice. To improve the point-spread function of the 2D-MRSI acquisition without any loss of SNR and time efficiency, a density-weighted concentric rings k-space trajectory (DW-CRT) sampling approach is used for the k-space acquisition trajectory of MRSI (2).Methods
All scans were acquired using a Siemens Prisma 3-Tesla (Siemens, Erlangen, Germany) whole-body MRI scanner and a 20-channel head array receive coil. B0 shimming was achieved using GRESHIM (3). The non-water-suppressed metabolite-cycling MRSI acquisition was achieved by utilizing two asymmetric narrow-transition-band adiabatic RF pulses with mirrored inversion profiles applied in alternate scans for the inversion of the upfield and downfield (relative to water) spectral ranges (4) before the semi-LASER localization with a gap of 9.6 ms. The semi-LASER (5) localization (TR = 1.5 s, TE = 68 ms), was used to excite whole brain slice with a 125 mm x 105 mm x 20 mm region centrally within the field of view (FOV= 240 mm x 240 mm). The Gaussian editing pulses of 65 Hz width were placed on the H3’ protons at 1.9 ppm in the “ON” acquisition, and at 9.0 ppm in the “OFF” acquisition. For DW-CRT, 64 points per ring were collected with an ADC bandwidth of 80 kHz, resulting in 512 temporal samples in an effective spectral bandwidth of 1250 Hz (Figure 1). To cover the 32x32 grid, 16 rings resulting in an individual voxel size of 1.125 mL (7.5 mm x 7.5 mm x 20 mm) were acquired for DW-CRT. The Hanning filter weighting function with an alpha value of 1 was used to compute radial sampling locations for the DW-CRT (2). For DW-CRT, four spatial interleaves (64 unique rings) were used to ensure that minimum field of view requirement was met, resulting in an acquisition duration of 384 s with metabolite cycling and mega editing. The number of averages was 2, corresponding to a total acquisition duration of 12.8 minutes. The feasibility of the whole brain slice with the proposed methods was tested on a healthy volunteer (male; 31 years old) after giving informed consent under an institutionally approved technical development protocol. All reconstructions were implemented in MATLAB using the non-uniform FFT (NUFFT) toolbox (6). Lipid contamination was removed during post‐processing using a lipid‐basis penalty algorithm (7).Results
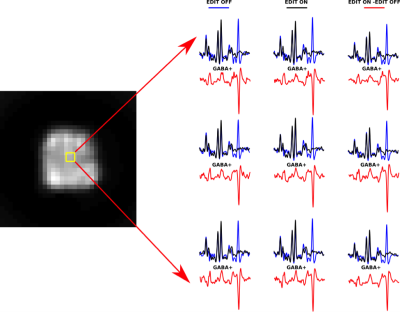
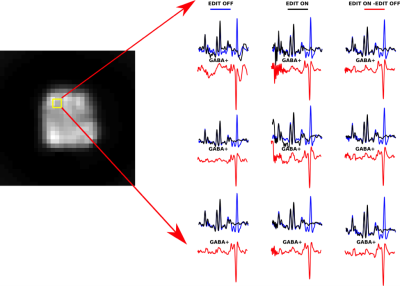
Figure 2 shows the non-water suppressed image of a whole brain slice as well as in-vivo edited spectra from the same slice of a healthy volunteer using metabolite cycling. After subtraction of the even MR spectra from the odd MR spectra, the edited in vivo MR spectra from 1.125 mL voxels showed well-defined GABA peaks at 3.0 ppm both in a central brain region (Figure 2) and a cortical brain region close to the lipid layer (Figure 3).Conclusion
We have developed and demonstrated a non-water suppressed MEGA semi-LASER MRSI sequence with DW-CRT that performs robustly at 3 Tesla, within a clinically feasible acquisition time of 13 min. The feasibility of the detection of GABA with the proposed method was shown in a healthy volunteer with a high in-plane resolution of 7.5 × 7.5 mm2. Experiments qualitatively demonstrate the advantage of DW-CRT in terms of its improved resolution and reduced contamination of spectra from neighboring voxels. Future work will focus on implementing the method for whole brain coverage with a reduced TR.Acknowledgements
This work was supported by NIH R01 ES020529References
1. Emir UE, Burns B, Chiew M, Jezzard P, Thomas MA. Non-water-suppressed short-echo-time magnetic resonance spectroscopic imaging using a concentric ring k-space trajectory. NMR Biomed 2017;30(7).
2. Chiew M, Jiang W, Burns B, Larson P, Steel A, Jezzard P, Albert Thomas M, Emir UE. Density-weighted concentric rings k-space trajectory for 1H magnetic resonance spectroscopic imaging at 7 T. NMR in Biomedicine:e3838-n/a.
3. Shah S, Kellman P, Greiser A, Weale P, Zuehlsdorff S, Jerecic R. Rapid Fieldmap Estimation for Cardiac Shimming. Proceedings 17th Scientific Meeting, International Society for Magnetic Resonance in Medicine. Honolulu 2009. p 565.
4. Dreher W, Leibfritz D. New method for the simultaneous detection of metabolites and water in localized in vivo 1H nuclear magnetic resonance spectroscopy. Magn Reson Med 2005;54(1):190-195.
5. Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. MAGMA 2008;21(1-2):95-101.
6. Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Transactions on Signal Processing 2003;51(2):560-574.
7. Bilgic B, Chatnuntawech I, Fan AP, Setsompop K, Cauley SF, Wald LL, Adalsteinsson E. Fast image reconstruction with L2-regularization. J Magn Reson Imaging 2014;40(1):181-191.
Figures