3950
Feature decomposition based examining tumor heterogenity on dynamic susceptibility contrast enhansed MRI data1Department of Radiology and Imaging Sciences, Emory University School of Medicine, Emory University, Atlanta, GA, United States, 2Long Hua Hospital, Guangdong, China, 3Department of Radiation Oncology, Emory University School of Medicine, Emory University, Atlanta, GA, United States
Synopsis
Dynamic susceptibility contrast-enhanced magnetic resonance imaging (DSC MRI) is widely used for studying blood perfusion in brain tumors. We report
PURPOSE Glioblastoma (GBM) is one of the most deadly cancers among adults.1 Dynamic susceptibility contrast-enhanced (DSC) magnetic resonance imaging (MRI) is a widely used imaging method to characterize the blood perfusion to the tumor tissue.2 Traditional analysis of DSC MRI data applies fixed models for analyzing signal time courses to derive the perfusion parameters despite of evidences that heterogeneous patterns of hemodynamic changes in different tumor regions.3 Since extracted information on tumor physiology is limited by the fixed model and unable to reveal the complexity of highly heterogeneous tissues in brain tumors, the present study aims to identify important signal time course features/patterns of DSC MRI profiles using a model-free approach with the assistance of artificial intelligence for feature decomposition.
METHOD DSC MRI was acquired from 14 patients with high-grade glioma and 8 patients with low-grade glioma (All cases, Age=45~70) on a 3T scanner (Siemens) using gradient echo single-shot echo planar imaging (TR/TE/FOV=45ms/2000ms/22cm, volume= 50 or 70, 25 axial slices, matrix=128×128). FSL software (http://fsl.fmrib.ox.ac.uk/fsl) was used for data pre-process which included noise reduction, image alignment, followed by skull stripping performed in the FLAIR image space using the Brain Extraction Tool provided by FSL. K-means clustering with silhouette criterion and 18-connected pixel connectivity information were used to segment tumor regions which were then spatially normalized into DSC MRI space and binarized as masks (inputs) to the different independent components (ICs) of signal time courses from the decompositions of DSC data using independent component analysis (ICA) implemented in the Melodic package (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/MELODIC). To select ICs, a defined criteria were established to filter out noise components based on the principle of the area of the curve (<5). The residual ICs with time courses have been found to exhibit heterogeneous patterns of hemodynamic changes in tumor regions. The numbers of selected ICs from the patients in different groups were used in the group comparison (high-grade glioma & low-grade glioma) using independent sample t-test with a threshold of p<0.05.
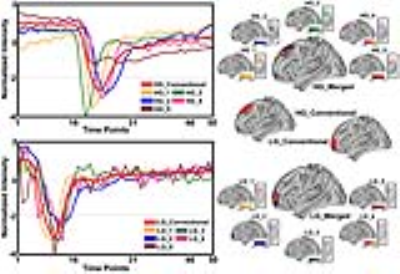
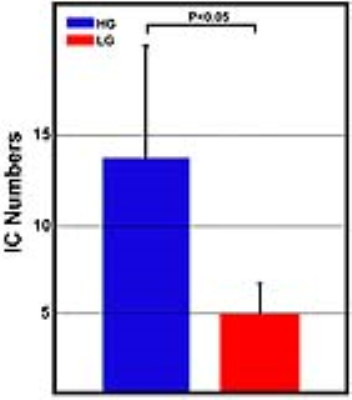
RESULTS Under the current selection criteria, the ICs of signal time courses shown in Figure 1 exhibited various patterns, not only including typical patterns captured in the conventional model-based DSC data analysis (e.g., IC HG_2, HG_4), but also atypical patterns (e.g., IC HG_5). Furthermore, these different patterns of signal profiles can be seen in the different regions of tumors (Figure 1), explicitly demonstrating spatially inhomogeneous blood perfusion properties in different tumor regions in both low and high-grade tumors. When we compared the numbers of heterogeneous patterns of time course changes in high-grade and low-grade gliomas (Figure 2), the numbers of ICs found in patients with high-grade glioma are significantly higher than those from patients with low-grade glioma (p<0.05) after filtering out noise components.
DISCUSSION AND CONCLUSION The heterogeneous characteristics of brain tumors were well known from analyzing the morphological images. However, the results of our current study revealed that the physiological properties such as blood perfusion are also heterogeneous within a tumor. The model-free and AI-based feature extraction allow for identifying and expanding more information from the perfusion MRI data comparing to the model based conventional DSC data analysis. Observed fewer numbers of ICs in low-grade glioma population in comparison with that in high-grade glioma not only suggests that tumors with lower grades are less heterogeneous comparing to the higher grade, which is consistent with early findings, but also proved the validity of the current approach of analyzing perfusion MRI data. Importantly, identifying and interrogating atypical perfusion MRI data time courses may allow us to investigate the tumor vascular features of those tumor tissues and regions and potentially develop new imaging markers for tumor progression and responses to the treatment.
Acknowledgements
No acknowledgement found.References
1. Stupp R et al., 2005. N Engl J Med. 352:987-96. 2. Lehmann P et al., 2010. Eur Neurol. 64:21-6. 3. Andriy Marusyk et al., 2010. Biochim Biophys Acta. 1805 (1): 105–117.
Figures