3921
Development of a Patient-specific Tumor Mold using MRI and 3D Printing Technology for Targeted Tissue Procurement and Radiomics Analysis of Renal Masses1Radiology, UT Southwestern Medical Center, Dallas, TX, United States, 2Urology, UT Southwestern Medical Center, Dallas, TX, United States, 3Pathology, UT Southwestern Medical Center, Dallas, TX, United States, 4Advanced Imaging Research Center, UT Southwestern Medical Center, Dallas, TX, United States
Synopsis
To implement a platform for co-localization of in vivo quantitative multi-parametric magnetic resonance imaging (mpMRI) features with ex vivo surgical specimens of patients with renal masses using patient-specific 3D-printed tumor molds, which may aid in targeted tissue procurement and
Introduction
Renal cell carcinoma (RCC) is a highly heterogeneous tumor.1 The potential of radiogenomics to differentiate among different subtypes of renal cell carcinoma has been recently illustrated.2, 3 Multi-parametric magnetic resonance imaging (mpMRI) protocols provide a unique opportunity to correlate objective imaging data with histologic and molecular alterations in the same tumor. However, image-tissue correlations are challenged by the complex anatomy, variability of location and size of renal tumor, and respiratory motion during MRI acquisitions. Three-dimensional (3D) printing has received much attention recently as such 3D models can help planning of surgical procedures and facilitate the education of patients and trainees.4-6 We expand the use of 3D models to explore its potential role in the field of radiomics. Thus, the purpose of our study was to generate an MRI-based patient-specific 3D mold for renal tumors and to assess our preliminary experience correlating MRI with histopathologic findings.Methods
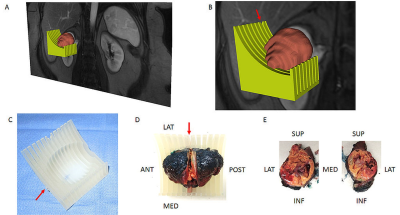
The institutional review board approved this Health Insurance Portability and Accountability Act-compliant prospective study. Written informed consent was obtained from all patients prior to mpMRI. All MRI examinations were performed on a 3T whole-body MRI system (Ingenia, Philips Healthcare, Best, The Netherlands) using a dStream anterior and posterior torso coils. Axial and coronal T2-weighted anatomic images were acquired followed by a coronal 2-dimensional (2D) arterial spin labelling (ASL) sequence, diffusion-weighted imaging (DWI), and dynamic-contrast enhanced (DCE)-MRI before and after the administration of a bolus 0.1 mmol/kg body weight of gadobutrol (Gadavist; Bayer Healthcare Pharmaceuticals, Wayne, NJ) at a rate of 2 mL/s followed by a 20 mL saline flush.7 We selected a set of coronal 3D post-contrast images from the DCE acquisition demonstrating a good contrast between the tumor and the renal parenchyma and transferred into 3D Slicer.8 We then generated a 3D model of the tumor using the model maker module in 3D Slicer and saved it in the standard tessellation language (STL) file format. A slicing guide template was created in the computer-aided design (CAD) software (SolidWorks; Dassault Systemes, France), which has notches corresponding to the anatomic locations of the MRI images (Fig. 1). Both the 3D tumor model and slicing guide STL models were imported into Netfabb (Netfabb GmbH, Parsberg, Germany). A Boolean operation was then performed in Netfabb, where the anatomic tumor model was subtracted from the slicing guide model, leaving a depression in the slicing guide corresponding to the exact size and shape of the tumor. The mold was labeled (i.e. anterior, posterior, superior, inferior) depending on the location of the primary tumor. Then, this customized, tumor-specific, slicing guide was printed using a 3D printer (Projet 3512HD, Rock Hall, SC) as a 3D mold (Fig. 1). After partial nephrectomy, the tumor was placed on the 3D mold and bivalved through the center slice notch to match the MRI plane. Targeted tissue samples were then obtained for radiogenomic analysis.Results
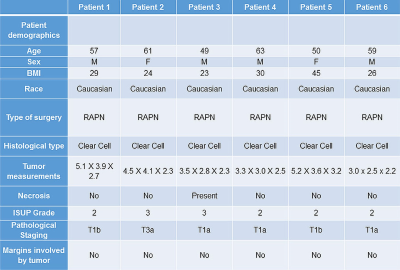
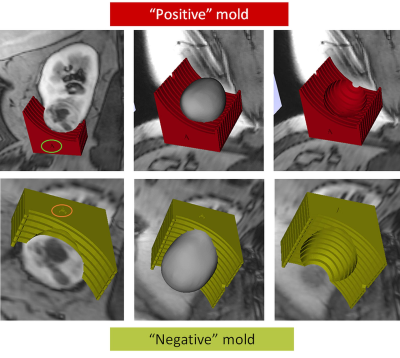
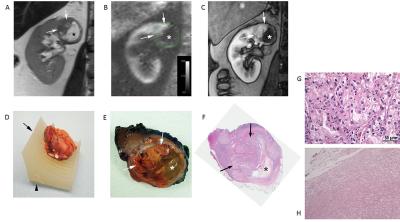
We generated patient-specific 3D molds in 6 patients who underwent an mpMRI examination. Patient demographics, tumor characteristics, and surgical data are presented in Table 1. For the first patient we created two 3D molds, one of the outer contour of tumor (positive mold) and one of the internal surface of the tumor (negative mold) where the tumor contacts the renal parenchyma (Fig. 2). Based on this case it became apparent that negative molds result in more precise fitting of the tumor within the 3D mold. This was primarily because the amount of retroperitoneal fat resected during a partial nephrectomy is more variable from patient to patient and interferes with the specimen sitting in the positive mold. In contrast, the inner contour of the tumor is more precisely resected during the enucleation in a partial nephrectomy. Accordingly, for the following 5 patients we only printed the negative 3D molds representing the inner contour of the tumor. Representative images of the correlation between MRI and gross specimen are presented in Fig. 3.Discusssion
Description about the use of 3D molds for MRI-histopathology correlations have been primarily limited to prostate specimens.9 To the best of our knowledge, this is the first report of the use of 3D-printed patient-specific molds to correlate MRI features with histopathology characteristics in the same kidney tumors. The combination of quantitative MRI techniques with the 3D patient-specific tumor mold offers a new opportunity for radiomic and radiogenomic analysis.Conclusion
We present a workflow for generating MRI-based patient-specific 3D molds of renal tumors that allow proper co-localization of MRI features in vivo with histopathologic characteristics in the same tumor and facilitates correlations with tissue-based analyses for radiomics and radiogenomic studies.Acknowledgements
This work was partially funded by grants NIH # P50CA196516 and R01 5RO1CA154475.References
1. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-892.
2. Shinagare AB, Vikram R, Jaffe C, et al. Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group. Abdom Imaging 2015;40:1684-1692.
3. Jamshidi N, Jonasch E, Zapala M, et al. The Radiogenomic Risk Score: Construction of a Prognostic Quantitative, Noninvasive Image-based Molecular Assay for Renal Cell Carcinoma. Radiology 2015;277:114-123.
4. Silberstein JL, Maddox MM, Dorsey P, Feibus A, Thomas R, Lee BR. Physical models of renal malignancies using standard cross-sectional imaging and 3-dimensional printers: a pilot study. Urology 2014;84:268-272
5. Sodian R, Schmauss D, Markert M, et al. Three-dimensional printing creates models for surgical planning of aortic valve replacement after previous coronary bypass grafting. Ann Thorac Surg 2008;85:2105-2108.
6. Lim KH, Loo ZY, Goldie SJ, Adams JW, McMenamin PG. Use of 3D printed models in medical education: A randomized control trial comparing 3D prints versus cadaveric materials for learning external cardiac anatomy. Anat Sci Educ 2016;9:213-221
7. Zhang Y, Kapur P, Yuan Q, et al. Tumor Vascularity in Renal Masses: Correlation of Arterial Spin-Labeled and Dynamic Contrast-Enhanced Magnetic Resonance Imaging Assessments. Clin Genitourin Cancer 2016;14:e25-36.
8. Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-1341.
9. Shah V, Pohida T, Turkbey B, et al. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum 2009;80:104301
Figures