3879
Minimum echo-time PRESS-localized proton observed carbon edited (POCE) magnetic resonance spectroscopy for rat brain imaging using simultaneous editing and localization pulses.1Biomedical Engineering, McGill University, Montreal, QC, Canada, 2McGill University, Montreal, QC, Canada, 3Douglas Hospital Brain Imaging Center, Montreal, QC, Canada, 4Yale University, New Haven, CT, United States
Synopsis
Dynamic Carbon-13 (13C) magnetic resonance spectroscopy (MRS) remains to be the only noninvasive method capable of measuring neuroenergetics and neurotransmitter cycling in the brain1. Proton observed carbon edited (POCE) MRS2 is an attractive alternative to direct 13C methods due to improved signal-to-noise-ratio (SNR). This study reports a PRESS localized POCE sequence utilizing simultaneous editing and localization (SEAL-PRESS), which allows the TE to be reduced to a theoretically optimal value of ~1/JHC (8.1ms, in this implementation). The sequence was validated in phantom and in a rat preparation, and demonstrated >17% improvement in 13C labeled metabolites relative to a 12.6-ms PRESS-POCE sequence.
Purpose
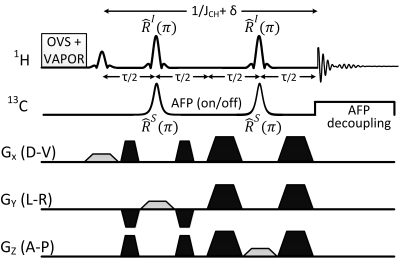
Indirect 13C magnetic resonance spectroscopy (MRS) by proton observed carbon editing (POCE) allows for the non-invasive investigation of neuroenergetics and neurotransmitter cycling in the brain in vivo1,2. The sensitivity of POCE-MRS can be enhanced through the use of short echo-time (TE) sequences, which minimizes T2 and homonuclear J-evolution related losses. Previous POCE-MRS implementations either use longer than optimal echo times due to sequence limitations, or short TE sequences based on image selected in vivo spectroscopy (ISIS) requiring multi-shot acquisitions for 3D localization. To that end, this work presents a novel single-shot PRESS-localized POCE-MRS sequence that involves the application of Simultaneous Editing And Localization pulses (SEAL-PRESS, see Fig 1), allowing the TE to be reduced to a theoretically optimal value of ~ 1/JHC (8.1-ms, in this implementation). The simultaneous application of editing and slice-selective refocusing pulses adds an additional consideration into the sequence design; namely, the normally broadband frequency selective carbon-editing pulses now become slice-selective.Methods
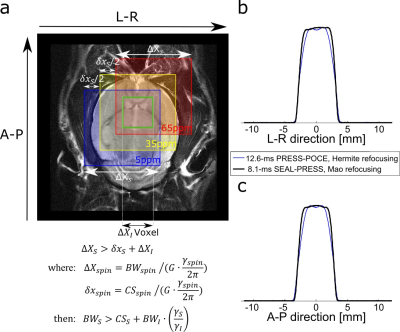
Editing efficiency of the SEAL-PRESS sequence was evaluated in simulation and subsequently implemented on a Bruker Biospec 70/30 horizontal bore preclinical scanner (120 mm diameter actively shielded gradient insert, 650 mT/m in 150 μs). Experiments were performed on two, adult male, Long Evans rats weighing 260-280 g. The tail vein of one rat was cannulated for a 0.85 M [1,6-13C2]-Glucose (Cambridge Isotope Laboratories Inc.) infusion, after which the animal was placed in the MRI scanner, under anesthesia (1-1.5% isoflurane). The 1H-[13C] coil setup consisted of a volumetric 1H transmitter, a 1H Rx-only surface coil, and a quadrature 13C transmit surface coil as previously described3. After a 2.5-hr dynamic in vivo 13C MRS scan session, the rat head was microwave-fixed (10-kW Muromachi Microwave Fixation System) and placed back in the MRI for further scanning. Animal preparation procedures were previously approved by McGill University’s animal research ethics committee and are in accordance with guidelines set by the council on animal care. The SEAL-PRESS sequence was implemented with the following parameters (TR = 4000 ms, TE = 8.1 ms). Both proton refocusing pulses RxI(π) were Mao (length = 1.3 ms, bandwidth = 4.8 kHz), and both carbon editing pulses RxS(π) were HS pulses [length = 1.3 ms, bandwidth (99% inversion efficiency) = 8.2 kHz, B2, max ~ 5 kHz]. The RxS(π) pulse bandwidth (BWS) required to cover the 13C chemical shift range (5-65 ppm, centered at 35 ppm, the resonance of [4-13C]-Glu) and within-voxel frequency shift, was calculated as illustrated in Fig 2a, since RxS(π) is applied during a slice-selective gradient. Thus the minimum required BWS of the HS pulse in this implementation was 5.7 kHz. Editing performance of the SEAL-PRESS sequence was compared against a previously described3 PRESS-POCE sequence (TR = 4000 ms, TE = 12.6 ms). An initial 58-mg bolus of [1,6-13C2]-Glc was infused during the first 15-seconds, and the rate was tapered off exponentially every 30-seconds to rapidly raise and maintain plasma glucose levels at ~10 mM as described previously4. Spectral fitting of POCE data was performed in LCModel using basis sets simulated in the FID-A toolkit.Results
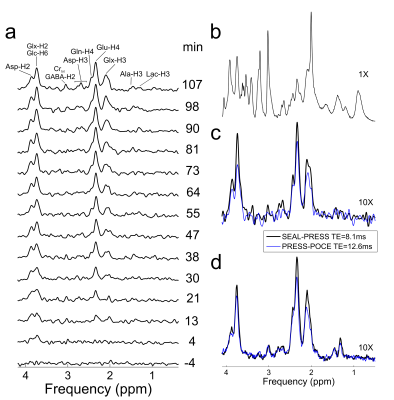
Fig 2b-c illustrates 1-D spatial profiles along both refocusing pulse directions (L-R, and A-P) obtained with the SEAL-PRESS and 12.6-ms PRESS-POCE sequences; sharper profiles were obtained for the SEAL-PRESS sequence due to the use of Mao pulses (without compromising sequence TE). Furthermore, near-uniform nutation in the region of interest is seen due to the homogenous B1+ field generated with the 1H volumetric transmitter4. Maximum editing efficiency of the SEAL-PRESS sequence was 95.5%, in comparison to ~ 96.3% obtained with the PRESS-POCE sequence with a classical editing scheme. Fig 3a illustrates time-resolved 1H-[13C] spectra obtained primarily encompassing the anterior cerebral cortex following the [1,6-13C2]-Glc infusion (100-μl voxel, 11-Hz water linewidth). Optimal sensitivity of spectra can be seen, with the last 8.5-min averaged acquisition (107-min) showing low-concentration 13C metabolites such as Lac-H2, Ala-H3, Asp-H3, and Asp-H2 in addition to the prominent Glu and Gln complexes. After 112-mins of time-resolved spectra, the [1,6-13C2]-Glc infusion was maintained to accommodate two 8.5-min in vivo scans each, with the SEAL-PRESS and 12.6-ms PRESS-POCE sequences for comparison (see Fig 3c) in an interleaved fashion (A=>B=>B=>A). Reduced T2 and homonuclear J-evolution related losses, as a result of reducing TE from 12.6-ms to 8.1-ms, improved 13C labeled metabolite signal by >17% (see Fig 3d).Conclusion
We present a single-shot PRESS-localized and
two-shot edited POCE MRS sequence with TE = 8.1ms. The sequence implementation
is straightforward, immune to scanner instabilities and subtraction artifacts,
while providing high sensitivity due to the short TE.Acknowledgements
This research was funded by NSERC (Grant No. RGPIN-2014-07072) awarded to JN.References
1. Rothman D et al. NMR Biomed, 2011;24(8):943-957.
2. Rothman D et al. et al. Proc Natl Acad Sci USA, 1985;82:1633-1637.
3. Kumaragamage C et al. MRM, 2017 [early view].
4. de Graaf R et al. NMR Biomed, 2011;24(8):958-972.
Figures