3877
Application of 3D-Dictionary Learning Compressed Sensing Reconstruction En Route to Isotropic Submillimeter Spatial Resolution Sodium (23Na) In Vivo MRI of the Human Eye at 7.0 Tesla1Berlin Ultrahigh Field Facility, Max Delbrueck Centrum, Berlin, Germany, 2Division of Medical Physics in Radiology, German Research Centre (DKFZ), Heidelberg, Germany, 3Institute of Radiology, Unviersity Hospital Erlangen, Erlangen, Germany, 4MRI.TOOLS GmbH, Berlin, Germany
Synopsis
Sodium ion (Na+) is a very important factor in the physiology of the human eye. However sodium (23Na) MRI is limited by its low sensitivity. Compressed sensing provides means to overcome this challenge. This work demonstrates the feasibility of high spatial resolution (1mm isotropic) 23Na in vivo MRI of the eye using a dedicated six-channel transceiver array in conjunction with a 3D dictionary learning compressed sensing algorithm. This approach showed distinct noise reduction along with substantial reduction in total acquisition time if benchmarked against conventional reconstruction employing standard gridding.
Introduction
Sodium ions (Na+) play a key role in the physiology of living cells. MRI is of proven value for probing Na+ content in vivo 1,2. Sodium (23Na) holds the second place in terms of NMR sensitivity among all nuclei present in biological tissues. Its accessible signal-to-noise ratio (SNR) is still around 3000 to 20.000 times lower than SNR of proton MRI. This limitation induces severe spatial resolution constraints and/or long acquisition times. To overcome this challenge, ultrahigh magnetic fields and ultrashort echo time imaging techniques are of substantial benefit. Compressed sensing (CS) based reconstruction methods provide an alternative for enhancing spatial resolution and for shortening scan times 3. This work examines the applicability 3D dictionary learning compressed sensing reconstruction (3D-DLCS) for isotropic (sub)millimeter spatial resolution 23Na in vivo MRI of the human eye at 7.0 Tesla.Methods
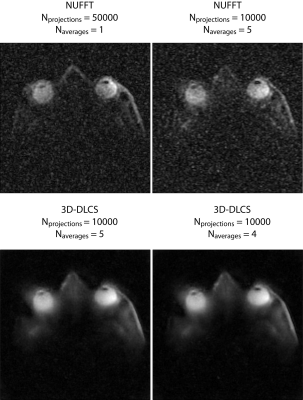
To acquire high-spatial resolution 23Na images of the human eye, we used a six-channel transceiver array that conforms very well to an average human head and a power divider which splits RF signal into six channels supporting equal amplitude and phase at all of the outputs. The multipurpose interface box (MRI.TOOLS GmbH, Berlin, Germany) consists of 16 transmit/receive switches (Stark Contrasts, Erlangen, Germany): 8 for 1H and 8 for 23Na resonant frequency at 7.0 T along with integrated low-noise preamplifiers (Stark Contrasts, Erlangen, Germany). 3D-density-adapted projection reconstruction was used for 23Na MRI data acquisition (TR = 17 ms, TE = 0.55 ms, FA = 41°). Proton MRI was conducted with a birdcage coil (Siemens, Erlangen, Germany) (Figure 1). Human imaging study was performed on a 7.0 Tesla whole-body system (Magnetom, Siemens Healthcare, Erlangen, Germany). 23Na MRI of the human eye was performed in one healthy, adult volunteer (sex: male; age = 26 years; BMI = 19.9 kg/m2). The data acquired with Nprojections = 50000, Naverages = 1, TA = 14.2 min were reconstructed with conventional gridding and applying Hamming filter (Figure 2). The data with Nprojections = 10000, Naverages = 5, TA = 14.2 min were additionally reconstructed with the 3D-DLCS algorithm (block size B = 6, dictionary size D = 500, weighting parameter for dictionary representation λ = 0.5, sample number Nsamp = 500000). Furthermore, one reconstruction was performed for a reduced dataset (Naverages = 4, TA = 11.3 min).Results
For sodium MRI of the eye two protocols were applied to achieve a nominal isotropic spatial resolution of 1 mm3 within an identical total acquisition time (TA) of 14 min 10 s: (A) Naverages = 1, Nprojections = 50000; (B) Naverages = 5, Nprojections = 10000. Both datasets (A) and (B) were reconstructed with a standard reconstruction method and were used as baseline images 4 (Figure 2). Dataset (B) was reconstructed with 3D dictionary learning compressed sensing (3D-DLCS) algorithm using 4 and 5 averages (Figure 2). Using only 4 averages instead of 5 does not lead to any drop in the performance of the 3D-DLCS reconstruction algorithm and the quality of the final image. This gain can be used to reduce total acquisition time for protocol (B) by almost 3 minutes. It is demonstrated that applying 3D 23Na dictionary learning compressed sensing reconstruction for 23Na in vivo MRI of the human eye at 7.0 Tesla preserves all of the details and leads to markedly reduced noise.Discussion and Conclusion
To summarize, 3D 23Na dictionary learning compressed sensing reconstruction is conceptually appealing for enhancing SNR in 23Na in vivo MRI of the human eye at 7.0 Tesla. This gain can be invested in reduction of total acquisition time or in spatial resolution enhancements. This improvement makes submillimeter 23Na MRI of the human eye at 7.0 Tesla feasible within acceptable scan times. This feasibility of high definition ocular 23Na MRI is of high relevance for precise diagnostics of various diseases of the eye including radiation planning and therapy monitoring of small ocular masses. Eye compartments, which are crucial in the context of sodium physiology, are very subtle (e.g. aqueous humor, lens). Resolving these subtle eye compartments requires high definition sodium MRI of the eye with submillimeter spatial resolution to obtain meaningful results from patient studies including ocular melanoma, glaucoma and cataract.Acknowledgements
No acknowledgement found.References
1. Madelin G and Regatte RR, JMRI, 2013.
2. Thulborn KR, Neuroimage, 2016.
3. Behl NGR, et al., MRM, 2015.
4. Wenz D, et al., ISMRM 2017.
Figures