3874
Voxel localization for sodium NMR triple-quantum signal; sequence design and test in agarose phantoms and in-vivo rat1Computer Assisted Clinical Medicine, Heidelberg University, Mannheim, Germany
Synopsis
The TQTPPI – sequence is a global spectroscopic method, which can be used to study the sodium metabolism of cells in-vitro with the foremost interest in the occurrence of the triple-quantum signal. This spectroscopic analysis could offer precise knowledge of sodium metabolic processes in pathology versus healthy tissue. To address clinical questions, we aim for transferring this method to in-vivo patient acquisitions where the need for a localization arises. Two different localization strategies, which preserve the quantum
Purpose and Introduction:
Previously the triple-quantum time proportional phase increment sequence (TQTPPI) was used to study the sodium metabolism of cells noninvasively.1,2 Of foremost interest is the occurrence of the triple-quantum signal (TQS), for slow motion sodium ions, e.g., bound to the sodium-potassium pump. This spectroscopic analysis could offer precise knowledge of sodium metabolic processes in pathology versus healthy tissue. To address clinical questions, we aim for transferring this method to in-vivo patient acquisitions, where the need for localization arises. We investigated two different strategies to localize the TQTPPI sequence preserving the quantum coherences and evaluated both sequences on agar-phantoms and in-vivo on a healthy rat.Subjects and Methods:
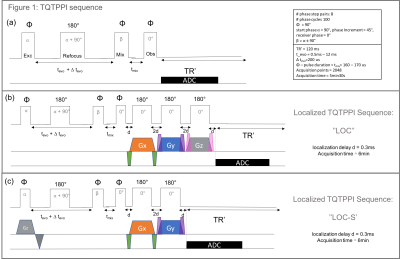
Sequence design: The global spectroscopy sequence (TQTPPI) uses triple quantum time proportional phase increments to study the 23Na – NMR spectrum, detecting single-quantum (SQ) and triple-quantum coherences (TQ) simultaneously.4 The TQTPPI sequence was extended with three extra 180° block pulses with simultaneous gradients ‘’LOC’’. Alternatively, we added a slice selection gradient in time with the first 90° excitation pulse of the TQTPPI sequence and only two additional 180° pulses ‘’LOC-S’’ (Figure1). Crusher gradients around the localizing gradients were implemented for both sequences. The three spatial directions were used for each crusher and a maximal strength of 15% of the localizing gradient strength was used.3 The additional pulses were added on purpose after the sequence block of TQTPPI to preserve the quantum coherences. Additionally, we investigated the effect of increased evolution time to optimize the normalized TQS in-vivo and in agar-phantom. TQS peak height normalized to its respective SQ peak height served as a first qualitative measure.
Data acquisition: Data were acquired on a 9.4T small-animal scanner (Bruker, Karlsruhe, Germany) using an in-house-built transmit/receive 23Na-surface-coil. An ultra-short echo time sequence was used for agarose phantom to obtain a sodium image for voxel positioning. For in-vivo rat measurement, we acquired a 1H T1 weighted gradient echo image with a volume transmit/receive coil, saving the isocenter position for the 23Na-surface-coil.
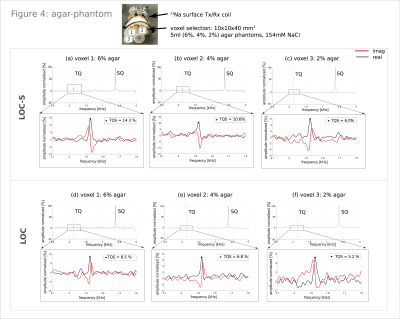
Phantoms/in-vivo: A 45ml agar-phantom, 134.75mM 23Na with 4% agarose used for initial sequence testing. Subsequently, we tested the localization in-vivo on a rat, scanned in the head region. Additionally, a phantom stack of 5ml-phantoms containing 6%, 4% and 2% agarose and 154mM 23Na was measured to investigate the possibility of tracing back agarose content based on TQS values.
Results:
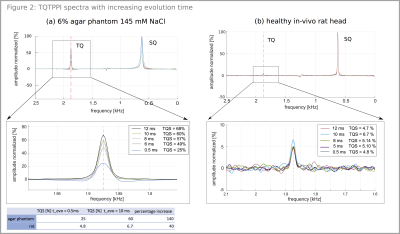
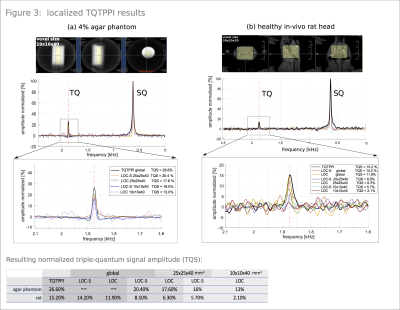
By increasing the evolution time from 0.5ms to 10ms, a TQS weighting was realized, facilitating localization (Figure2). For a healthy in-vivo rat, we observed an increase of 40% compared to the initial TQS, for agar-phantom a rise of 139%. Localization measurements preserved the quantum coherences for both sequences. Agar-phantom result and in-vivo rat result is depicted in Figure3(a,b) respectively. Compared to the global TQTPPI sequence we recorded a TQS drop of 1% and 3.3% for LOC-S and LOC respectively when choosing a global voxel size of 100x100x100mm3. Resulting normalized TQS values can be found in Figure3. TQS decreased with smaller voxel sizes, a voxel size of 10x10x40mm3 is 16% in volume of the voxel size 25x25x40mm3 and TQS dropped by 22% for LOC-S and by 26% for LOC(Figure3). Figure4 illustrates results of the agar-phantom-stack with a voxel size of 10x10x40 mm3. The resulting spectra for the 6%, 4% and 2% agar tubes show a decreasing TQS for both sequences (Figure4). Therefore, it was possible to distinguish the tubes based on their relative result in TQS with both sequences.Discussion:
With current settings, LOC-S triple-quantum signal was higher for equal voxel size and material than for LOC sequence. However, the latter sequence takes longer before data readout due to the additional 180° pulse which leads to signal drop. Further optimization of the crusher gradients and pulse shapes could enable smaller voxel sizes. Increasing the evolution time enhanced the TQS due to the difference in relaxation time of single-quantum and triple-quantum signal. This effect could be phantom dependent as we measured a difference of 2ms for the optimum evolution time. Especially for localization, where low TQS is problematic, it is useful to measure at the optimum time point. The exact time point could be further evaluated in a simulation.Conclusion:
We successfully designed two types of localization for the TQTPPI sequence without destroying quantum coherences. Localization performed well in-vitro and in-vivo for voxel sizes of 10x10x40mm3. In-vitro results further showed that a varying agar concentration could be distinguished based on the triple-quantum signal with both sequences. In the next step, we want to transfer the sequences to a human system enabling the study of the sodium metabolism in healthy tissue and pathology.Acknowledgements
Generous support in animal handling was received from Felix Hörner of Zentralinstitut für Seelische Gesundheit, Mannheim, Germany.References
1. Schepkin, V. D., Neubauer, A., Nagel, A. M., & Budinger, T. F. (2017). Comparison of potassium and sodium binding in vivo and in agarose samples using TQTPPI pulse sequence. Journal of Magnetic Resonance, 277, 162–168.
2. Neubauer, A., Nies, C., Schepkin, V. D., Hu, R., Malzacher, M., Chacón-Caldera, J., Schad, L. R. (2017). Tracking protein function with sodium multi quantum spectroscopy in a 3D-tissue culture based on microcavity arrays. Scientific Reports, 7(1), 3943.
3. Bernstein, M. A., King, K. F., & Zhou, X. J. (2004). Handbook of MRI Pulse Sequences. Elsevier Inc.
Figures