3862
High resolution 3D EPSI Spectroscopic Imaging compared with Spectral Localization by Imaging (SLIM)1Hoglund Brain Imaging Center, University of Kansas Medical Center, Kansas City, KS, United States, 2Department of Bioengineering, University of Kansas, Lawrence, KS, United States, 3Department of Molecular & Integrative Physiology, University of Kansas Medical Center, Kansas City, KS, United States, 4Department of Neurology, University of Kansas Medical Center, Kansas City, KS, United States
Synopsis
Quantitative measurements of metabolites in important structures in the human brain are challenging to acquire using conventional spectroscopic imaging methods. In this study, we compared regional metabolite concentrations obtained from high resolution 3D EPSI MRSI data using two different methods: spatial averaging inside regions of interest using the MIDAS software, and Spectral Localization by Imaging (SLIM)-based MRS. Deep brain structures were studied and compared using the two methods. The quantitative outcomes of 3D EPSI and SLIM were comparable and SLIM could provide metabolite concentrations using shorter scan times by reducing the required number of voxel acquisitions.
Introduction
High resolution 3D EPSI offers to overcome resolution barriers in spectroscopic imaging of the brain. However, the acquisition time of EPSI is very long, in the range of 20 minutes per scan. The theory of Spectral Localization by Imaging (SLIM) provides a basis for reconstruction of the spectra of anatomical compartments in under-sampled conditions, by taking advantage of high resolution prior anatomical segmentation information from MRI. SLIM could thus utilize fewer k-space acquisitions to achieve performance comparable with region-of-interest (ROI) averaging of 3D EPSI. The purpose of this study was to evaluate MRSI reconstructions acquired by 3D EPSI, using both full k-space ROI averaging and SLIM with reduced k-space sizes.Background and Theory
Echo planar spectroscopic imaging (EPSI)
can be used to acquire full 3D k-space data with coverage of the whole
brain. However, the scan time of EPSI is
very long, due to the large number of phase encoding acquisitions. Spectral Localization by Imaging (SLIM)
provides a framework to incorporate high resolution prior information from MRI,
enabling the reconstruction of chemical shift spectra of distinct anatomical
compartments [1]. The SLIM problem is expressed by a matrix equation s = G*c,
where s is a vector of phase encoded signals, G is a geometry matrix derived
from the compartment information and phase encoding vectors, and c is a vector
of the compartment coefficients. Solving
the system of SLIM requires a number of phase encodes larger than the number of
compartments, and generally a greater number of phase encodes improves the resilience
of SLIM reconstruction in the presence of compartmental inhomogeneities. Considering the large k-space coverage offered
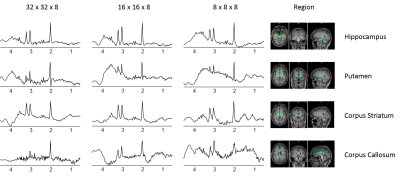
by EPSI, we aimed to investigate the performance of SLIM reconstruction with
different k-space sizes including localized reconstructions from significantly
reduced k-space.
Methods
Four healthy control subjects were consented
according to institutional review board approved protocols. Scans were performed on a 3 T scanner (Skyra,
Siemens) with a 16 channel head receiver coil. 1H MRSI was acquired using a 3D EPSI sequence
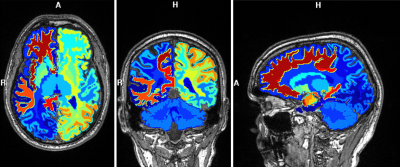
[2] (TE/TR=3980/200000 ms). Parcellation of gray and white matter was obtained
from MPRAGE images using FreeSurfer [3] to provide regional anatomical compartments
for MIDAS and SLIM. EPSI reconstruction was performed by Metabolite Imaging and Data Analysis
Software (MIDAS). Nine deep gray and
white matter structures were selected and processed through MIDAS ROI averaging and
SLIM reconstructions. Metabolite estimation and spectral fitting
was processed by LCModel [4].
Results and Discussion
The acquisition time for 3D EPSI is around 20 minutes, making clinical application challenging. The SLIM theory promises to allow signal localization of compartments using much fewer k-space acquisitions, by taking advantage of prior information from high resolution MRI to reconstruct the spectra of anatomical compartments. The results of this study showed that localization of anatomical compartment spectra using SLIM could be retrieved from identically shaped structures used in MIDAS, with greatly reduced number of phase encoding acquisitions. This shows promise of using advanced reconstruction by SLIM and related techniques to improve the performance of 3D MRSI applications for quantitative spectroscopy of the human brain.Acknowledgements
The Hoglund Brain Imaging Center is supported by the NIH (S10RR029577) and the Hoglund Family Foundation.References
1. Hu X, et al., SLIM: spectral localization by imaging. Magn Reson Med, 8:314-322, 1988
2. Maudsley A.A., et al., Comprehensive processing, display, and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 19:492-503, 2006
3. Dale, A.M., et al., Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9:179-194, 1999
4. Provencher S.W., et al., Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn. Reson. Med. 30:672–679, 1993
Figures