3854
HOPE (Half-intensity with macrOmolecule-suPprEssion): Ultra-short TE MRS without macromolecules on 3 T, 4 T and 9.4 T1McLean Hospital; Harvard Medical School, Belmont, MA, United States, 2National Institute on Drug Abuse, Baltimore, MD, United States
Synopsis
The strong and overlapping macromolecule (MM) signals remains as one of the major technical challenges for metabolites quantification using ultra-short TE MRS. HOPE (Half-intensity with macrOmolecule-suPprEssion), a simple but effective MM suppression MRS method based on SPECIAL, was proposed and tested on human 3T, 4T and animal 9.4T. HOPE has no additional pulse or cycling compared to SPECIAL but with the same short TE. With the similar SNR level as STEAM, HOPE achieves additional benefit with substantial suppression of MM signals and more accurate quantifications of MM overlapped metabolites such as lactate and GABA.
Introduction
It has been demonstrated that the ultra-short spin echo time (TE) proton MRS methods greatly enhanced the reliabilities detecting smaller and frequency overlapping compounds in the brains of both rodents (1,2) and human (3,4) because J modulation of scalar coupled metabolites are greatly reduced. However, the strong and overlapping macromolecule (MM) signals remains as the major technical challenge for metabolites quantification using short TE MRS (5). MM resonances have been reported to vary with age and cerebral location (6,7), as well as composition of gray/white matter (7,8). Large inter-subject variance of MM has also been reported (9). Mathematical (10) and experimental (11,12) estimates of MM are the two most widely used methods to account for the MM contributions, but there are still difficulties to address individual MM characteristics. In this study, HOPE (Half-intensity with macrOmolecule-suPprEssion), an ultra-short TE MRS sequence based on SPECIAL (13,14) to achieve effective MM suppression, was proposed (15) and implemented on 3 T, 4 T and 9.4 T to demonstrate its capability to detect MM overlapped compounds such as lactate (Lac) and GABA.Methods
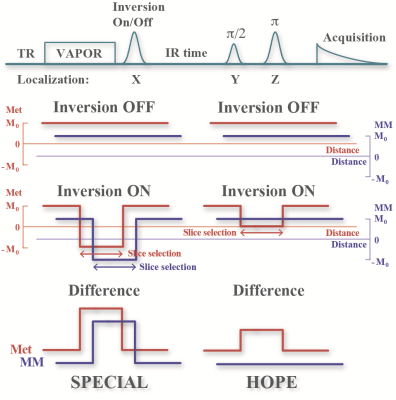
The mechanism of HOPE is demonstrated in Fig. 1: a delay is inserted after the inversion localization pulse of SPECIAL. With a well-adjusted delay time, the MM resonances are mostly subtracted out in the difference spectrum, with metabolite peaks remains with approximately half intensity because MMs’ T1 are much shorter than metabolites. No additional pulse or cycling is needed compared to SPECIAL.
The feasibility of HOPE was tested on the Siemens 3 T Trio human scanner with a 32-channel birdcage coil, the Agilent 4 T human scanner with a quadrature surface coil, and Bruker 9.4 T animal scanner with a volume coil as transmitter and surface coil as receiver. The TE on 3 T, 4 T and 9.4 T were 8 ms,10 ms and 6.5 ms respectively. 5 and 4 healthy subjects went through the scan on 3 T and 4 T scanners respectively, with the MRS voxel on the anterior cingulate cortex (ACC) region (30 x 20 x 30 mm3). 6 rats were scanned on the 9.4 T animal scanner with MRS voxel on the prefrontal cortex (3 x 5 x 4 mm3). On all scanners 256 averages were acquired with TR = 3 s for the water suppressed spectrum and 16 averages for water spectrum. Average-by-average frequency and phase corrections were performed in the matlab based MRS toolkit FID-A (16). The eddy current corrections and fitting quantification was performed in LCModel (10) with the simulated macromolecule peaks accounting for the baseline.
Results and discussions
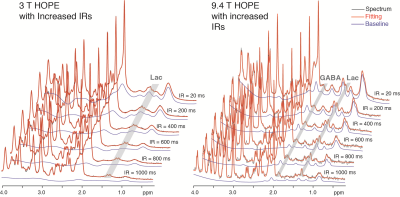
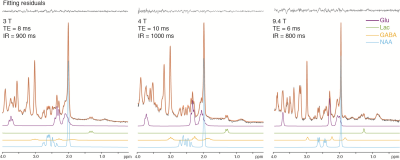
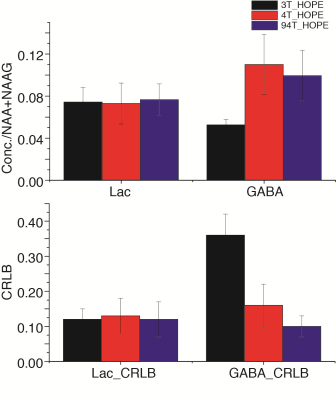
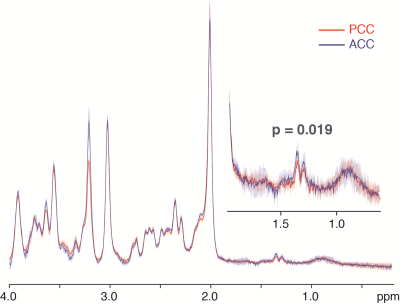
A series of inversion-recovery times (IRs) from 20 ms (regular SPECIAL) to 1 s were tested and the representative spectra on 3 T and 9.4 T were shown in Fig. 2 together with their baselines. MM signals gradually reduced and the baseline became smoother as IR increased, while the MM overlapping metabolites such as Lac and GABA became more resolved. A longer IR achieves better MM suppression however with lower SNR. With balancing MM suppression and SNR, IRs of 900 ms, 1000 ms and 800 ms were used for the scans on 3 T, 4 T human scanners and 9.4 T animal scanner respectively (Fig. 3). Lac doublet at 1.3 ppm was much better resolved with MM suppression on all field strengths. With suppression of MM near 2.0 ppm, the GABA resonances at 1.9 ppm were less overlapped and increasingly resolved as the field strength increases. The LCModel quantification results in Fig. 4 follow the same trend: the GABA concentrations measured with 4.0 T and 9.4 T were similar while the CRLB reduced as the static magnetic field increased. On the other hand, Lac measurements on different platforms were very consistent. With a similar measurement of PCC on 3 T of the same subjects, it was interesting to observe a significantly lower Lac level compared to ACC (Fig. 5, N=5, paired t-test) and the result is pending with further verification. NAA+NAAG was used as reference for all results above.Conclusion
HOPE, a simple but effective MM suppression MRS sequence with ultra-short TE was proposed and tested on a variety of magnetic field strengths. With the same SNR level as STEAM, HOPE achieves additional benefit with substantial suppression of MM signals and more accurate quantifications of MM overlapped metabolites. With better spectral resolution and higher SNR on human 7 T, it would be very promising to resolve all proton measurable metabolites without MM contaminations in one single scan.Acknowledgements
No acknowledgement found.References
1. Pfeuffer J, Tkac I, Provencher SW, Gruetter R. Toward an in vivo neurochemical profile: Quantification of 18 metabolites in short-echo-time H-1 NMR spectra of the rat brain. Journal of Magnetic Resonance 1999;141(1):104-120.
2. Mlynarik V, Cudalbu C, Xin L, Gruetter R. 1H NMR spectroscopy of rat brain in vivo at 14.1Tesla: improvements in quantification of the neurochemical profile. J Magn Reson 2008;194(2):163-168.
3. Tkac I, Andersen P, Adriany G, Merkle H, Ugurbil K, Gruetter R. In vivo H-1 NMR spectroscopy of the human brain at 7 T. Magnetic Resonance in Medicine 2001;46(3):451-456.
4. Deelchand DK, Van de Moortele PF, Adriany G, Iltis I, Andersen P, Strupp JP, Vaughan JT, Ugurbil K, Henry PG. In vivo 1H NMR spectroscopy of the human brain at 9.4 T: initial results. J Magn Reson 2010;206(1):74-80.
5. Cudalbu C, Mlynarik V, Gruetter R. Handling macromolecule signals in the quantification of the neurochemical profile. Journal of Alzheimer's disease : JAD 2012;31 Suppl 3:S101-115.
6. Hofmann L, Slotboom J, Boesch C, Kreis R. Characterization of the macromolecule baseline in localized (1)H-MR spectra of human brain. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2001;46(5):855-863.
7. Mader I, Seeger U, Weissert R, Klose U, Naegele T, Melms A, Grodd W. Proton MR spectroscopy with metabolite-nulling reveals elevated macromolecules in acute multiple sclerosis. Brain 2001;124(Pt 5):953-961.
8. Benoît S, Lijing X, Rolf G. Is the macromolecule signal tissue‐specific in healthy human brain? A 1H MRS study at 7 tesla in the occipital lobe. Magnetic Resonance in Medicine 2014;72(4):934-940.
9. Chong DG, Kreis R, Bolliger CS, Boesch C, Slotboom J. Two-dimensional linear-combination model fitting of magnetic resonance spectra to define the macromolecule baseline using FiTAID, a Fitting Tool for Arrays of Interrelated Datasets. Magma 2011;24(3):147-164.
10. Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed 2001;14(4):260-264.
11. de Graaf RA, Brown PB, McIntyre S, Nixon TW, Behar KL, Rothman DL. High magnetic field water and metabolite proton T1 and T2 relaxation in rat brain in vivo. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2006;56(2):386-394.
12. Kunz N, Cudalbu C, Mlynarik V, Huppi PS, Sizonenko SV, Gruetter R. Diffusion-Weighted Spectroscopy: A Novel Approach to Determine Macromolecule Resonances in Short-Echo Time H-1-MRS. Magnetic Resonance in Medicine 2010;64(4):939-946.
13. Mlynarik V, Gambarota G, Frenkel H, Gruetter R. Localized short-echo-time proton MR spectroscopy with full signal-intensity acquisition. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2006;56(5):965-970.
14. Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, Gruetter R. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2009;61(6):1279-1285.
15. Chen X, Rowland LM, Yang Y. In vivo short spin-echo 1H MR spectroscopy with macromolecule suppression. 2011; Montreal. p 3419.
16. Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)-An open source, MATLAB-based toolkit. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine 2017;77(1):23-33.
Figures