3853
Spatially localized pure shift 1H MRS for biological tissues at 7 T1Department of Electronic Science, Xiamen University, Xiamen, China
Synopsis
Proton magnetic resonance spectroscopy
(1H MRS) presents an effective tool for in vivo studies on biological tissues by the non-invasive detection manner. However, due to limited proton
chemical shift range and extensive J coupling splittings,
spectral congestions or even overlapping are generally encountered in resulting
1D 1H MRS acquired by routine STimulated Echo
Acquisition Mode (STEAM) and Point resolved spectroscopy (PRESS) experiments. In this report, we presents a previously-unreported
MAS approach to obtain spatially localized
1D pure shift spectra with spectral simplification, potentially useful for studies on biological
samples.
Introduction
Proton magnetic resonance spectroscopy (1H MRS) has emerged as a powerful tool for analyzing molecular structures and compositions, especially providing metabolite information complementing the insight delivered by MRI for in vivo studies on biological tissues.1,2 However, due to limited proton chemical shift range (~10 ppm) and extensive J coupling splittings, spectral congestion is generally observed in resulting 1D spectra acquired by routine MRS techniques, STimulated Echo Acquisition Mode (STEAM) and Point resolved spectroscopy (PRESS). A NMR technique, “pure shift spectroscopy”, provides a feasible way for removing J coupling effects and enhancing spectral resolution with only chemical shift present.3,4 In this report, a MRS approach based on the pure shift module, PSYCHE (pure shift yielded by chirp excitation), is introduced to obtain spatially localized 1D spectra with spectral simplification and resolution enhancement.Methods
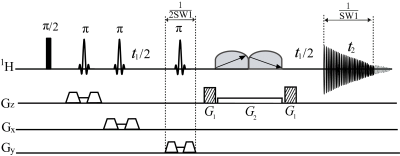
The proposed pure shift MRS sequence is shown in Fig. 1. This sequence is designed based on the concatenation of a localization module and a PSYCHE module. The localization module, consisting of three slice-selective refocusing π RF pulses along with slice-selective gradients and crusher gradients, is used for volume selection. The PSYCHE module, consisting of an indirect evolution period t1 and two frequency-swept chirp pulses β matching with a simultaneous weak gradient G2 and a pair of coherence selection gradients G1, generates the desired pure shift signals. Acquired data in the t2 period contains the chunk data of 1/SW1 interval for each t1 increment, and a 1D data without J coupling effects can be reconstructed after the concatenation of all chunked data through t1 increments. Thus, the spatially localized 1D pure shift spectrum is obtained after direct Fourier transform.
All experiments were performed on a Varian (Palo Alto, CA, USA) 7 T small animal magnetic resonance scanner at 293 K. A phantom built with two plastic bottles, filled with 1 M propionate (Prop) aqueous solution and 1 M γ-aminobutyric acid (GABA) aqueous solution, was used to demonstrate the performance of the pure shift MRS method. In addition, this method was also applied to an intact pig brain tissue to show its applicability on biological tissues.
Results
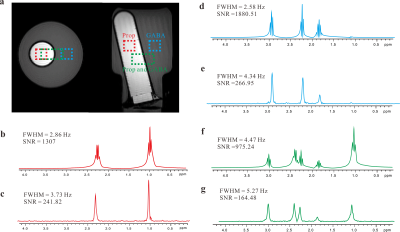
The comparison results on the phantom sample acquired from standard PRESS sequence and pure shift MRS sequence are shown in Fig. 2. Figure 2a shows axial and coronal spin-echo images of the sample, displaying three selected voxels, i.e. 5×5×5 mm3 for the GABA component (blue), 5×5×5 mm3 for the Prop component (red), and 5×10×5 mm3 for both two components (green). Multiplet peaks caused by J coupling splittings are observed in 1D PRESS spectra (Fig. 2b, d, and f). In contrast, the J coupling effect is suppressed and all multiplets are collapsed to singlets for each chemical shift size in resulting spectra obtained by the pure shift MRS approach (Fig. 2c, e, and g). The calculated signal to noise rati (SNR) and full width at half-maximum (FWHM) are given in PRESS spectra and pure shift MRS spectra for comparison.
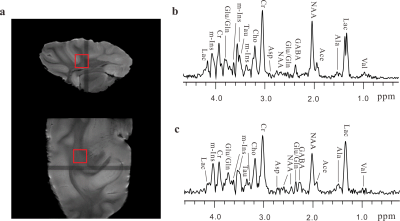
The experimental results on the intact pig brain tissue are shown in Fig. 3. The localized voxel with the size of 6×6×6 mm3 is marked in spin-echo images of the tissue (Fig. 3a). Figure 3b shows a standard 1D PRESS spectrum acquired from the selected voxel of pig brain tissues and most metabolites are assigned. The localized pure shift MRS spectrum from the same voxel is given in Fig. 3b. The localized pure shift spectrum can yield much better spectral resolution satisfactory for metabolite analyses. Most metabolites observed in the standard PRESS spectrum (Fig. 3b) can be clearly obtained in the localized pure shift spectrum (Fig. 3c).
Discussion
The original 1D PSYCHE provides a promising manner for simplifying complex spectral information by removing J coupling effects. Herein, we exploit this pure shift module to MRS applications with numerous metabolites, aiming for spectral simplification and spectral resolution enhancement. Experiments on the phantom and the intact pig brain tissues shows the applicability of the proposed pure shift MRS approach on spatially localized pure shift measurements. Due to the intrinsic signal penalty caused by the PSYCHE element, the sensitivity of pure shift MRS spectra is generally lower than that in standard PRESS spectra.Conclusion
Here, we present a previously-unreported MAS approach to obtain localized 1D pure shift spectra. Compared to the conventional MRS approaches, multiplets are collapsed to singlets by the proposed MRS method, potentially useful for metabolite analyses in biological samples containing numerous metabolite compositions.Acknowledgements
This work was partially supported by the NNSF of China under Grants 11675135 and 21327001.References
1. Pouwels PJW, Frahm J. Regional metabolite concentrations in human brain as determined by quantitative localized proton MRS. Magn. Reson. Med. 1998; 39(1):53-60.
2. Thomas MA, Lipnick S, Velan SS, et al. Investigation of breast cancer using two-dimensional MRS. NMR Biomed. 2009; 22(1):77-91.
3. Zangger K, Sterk H. Homonuclear broadband-decoupled NMR spectra. J. Magn. Reson. 1997; 124(2):486-489.
4. Foroozandeh M, Adams RW, Meharry NJ, et al. Ultrahigh-Resolution NMR Spectroscopy. Angew. Chem. Int. Edit. 2014; 53(27):6990-6992.
Figures